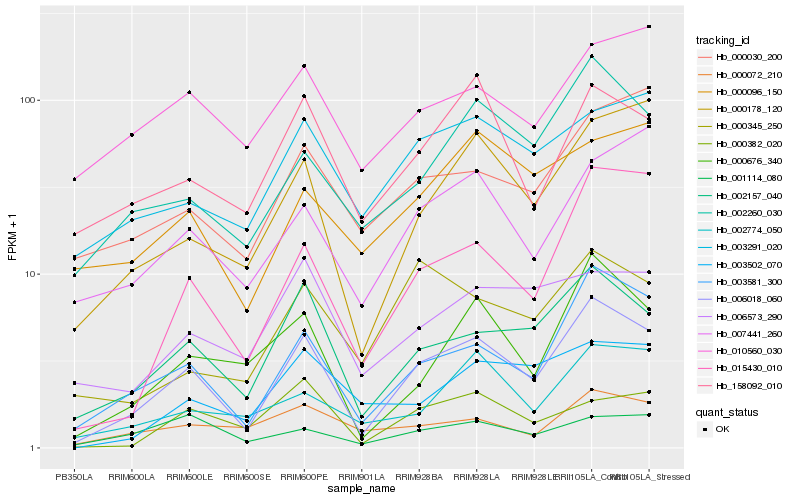
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006018_060 |
0.0 |
- |
- |
Zinc finger SWIM domain-containing protein 7 isoform 1 [Theobroma cacao] |
| 2 |
Hb_003581_300 |
0.1155825577 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002157_040 |
0.1319190455 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 4 |
Hb_015430_010 |
0.1603830261 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 5 |
Hb_000676_340 |
0.1630490536 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000178_120 |
0.165331295 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 7 |
Hb_002260_030 |
0.1672912791 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 8 |
Hb_158092_010 |
0.1893535405 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 9 |
Hb_002774_050 |
0.1915012481 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 10 |
Hb_003291_020 |
0.1916756506 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 11 |
Hb_007441_260 |
0.1935808794 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_003502_070 |
0.1966508811 |
- |
- |
hypothetical protein CISIN_1g0405401mg, partial [Citrus sinensis] |
| 13 |
Hb_000072_210 |
0.1967456766 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 14 |
Hb_000382_020 |
0.1981374577 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 15 |
Hb_006573_290 |
0.1998610476 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 16 |
Hb_000030_200 |
0.2024936731 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 17 |
Hb_001114_080 |
0.2026188264 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 18 |
Hb_000345_250 |
0.2031024569 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 19 |
Hb_000096_150 |
0.2031196724 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 20 |
Hb_010560_030 |
0.2032293838 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |