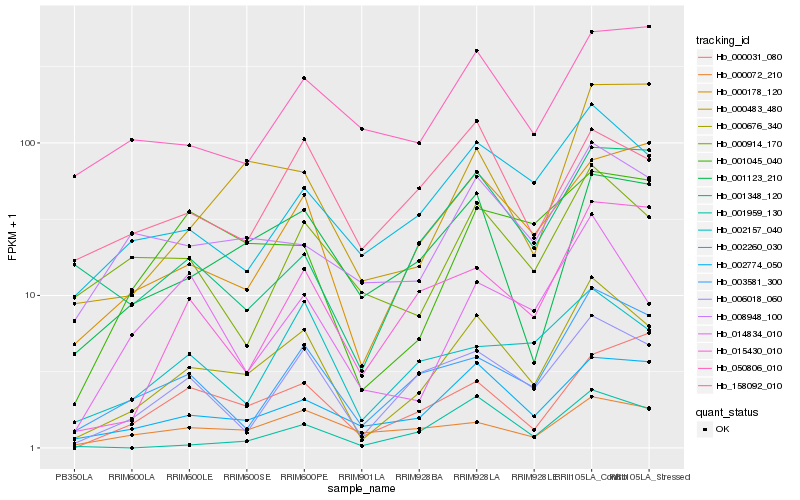
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_340 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_006018_060 |
0.1630490536 |
- |
- |
Zinc finger SWIM domain-containing protein 7 isoform 1 [Theobroma cacao] |
| 3 |
Hb_002774_050 |
0.1681197751 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 4 |
Hb_002260_030 |
0.1781274313 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 5 |
Hb_003581_300 |
0.1801165843 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001123_210 |
0.1850715814 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 7 |
Hb_000178_120 |
0.1871740237 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 8 |
Hb_008948_100 |
0.1929041763 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 9 |
Hb_002157_040 |
0.1979578729 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 10 |
Hb_001045_040 |
0.2015237673 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 2 [Jatropha curcas] |
| 11 |
Hb_000072_210 |
0.2039603142 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 12 |
Hb_158092_010 |
0.2049411774 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 13 |
Hb_014834_010 |
0.2056141765 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4 [Jatropha curcas] |
| 14 |
Hb_015430_010 |
0.2083371881 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 15 |
Hb_000914_170 |
0.2097886153 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 16 |
Hb_001959_130 |
0.2101206936 |
- |
- |
transporter, putative [Ricinus communis] |
| 17 |
Hb_000483_480 |
0.2185233245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000031_080 |
0.2205884194 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 19 |
Hb_050806_010 |
0.2214606401 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 20 |
Hb_001348_120 |
0.2272753501 |
- |
- |
Phloem protein 2-B10, putative [Theobroma cacao] |