| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
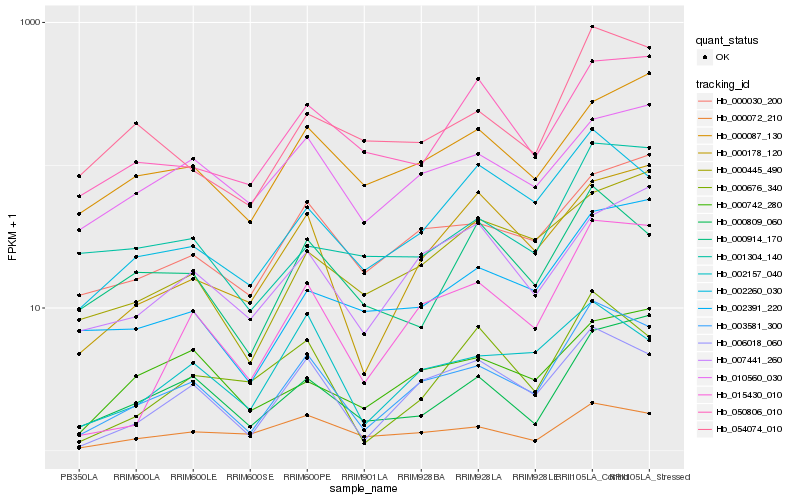
Hb_003581_300 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006018_060 |
0.1155825577 |
- |
- |
Zinc finger SWIM domain-containing protein 7 isoform 1 [Theobroma cacao] |
| 3 |
Hb_015430_010 |
0.1317658831 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 4 |
Hb_054074_010 |
0.1636877348 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 5 |
Hb_002260_030 |
0.1638329578 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 6 |
Hb_002157_040 |
0.1666701123 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 7 |
Hb_000030_200 |
0.1669829004 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 8 |
Hb_000178_120 |
0.1701189274 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 9 |
Hb_000809_060 |
0.1740711543 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 10 |
Hb_002391_220 |
0.1793350566 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 11 |
Hb_000676_340 |
0.1801165843 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000072_210 |
0.183794011 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g38730 [Jatropha curcas] |
| 13 |
Hb_001304_140 |
0.1844404789 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 14 |
Hb_000445_490 |
0.1854667485 |
- |
- |
- |
| 15 |
Hb_000742_280 |
0.1868717942 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_007441_260 |
0.1879348016 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000914_170 |
0.1890454257 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 18 |
Hb_000087_130 |
0.1892333251 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 19 |
Hb_050806_010 |
0.1892572225 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 20 |
Hb_010560_030 |
0.1897338817 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |