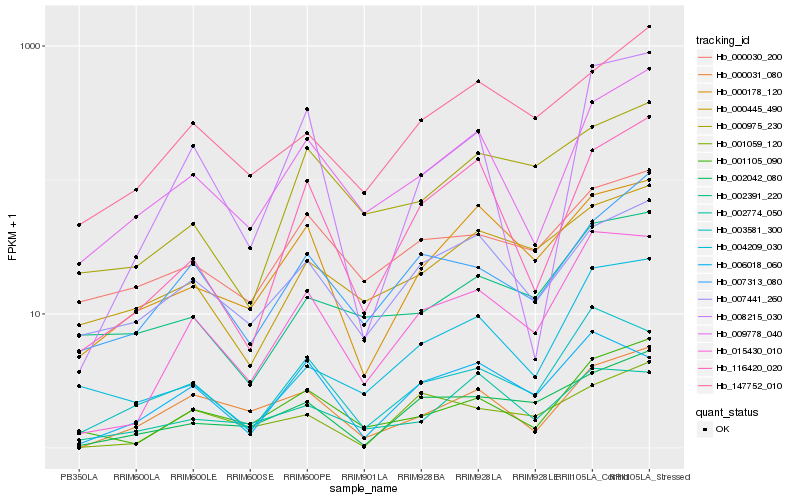
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015430_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 2 |
Hb_003581_300 |
0.1317658831 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001105_090 |
0.1475216877 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006018_060 |
0.1603830261 |
- |
- |
Zinc finger SWIM domain-containing protein 7 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000178_120 |
0.1646515562 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 6 |
Hb_002042_080 |
0.167380476 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 7 |
Hb_009778_040 |
0.1716266068 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 8 |
Hb_116420_020 |
0.1752871051 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 9 |
Hb_008215_030 |
0.1811781366 |
- |
- |
hypothetical protein 3 [Hevea brasiliensis] |
| 10 |
Hb_004209_030 |
0.1817942961 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000031_080 |
0.1829028732 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 12 |
Hb_000975_230 |
0.1843184766 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |
| 13 |
Hb_007441_260 |
0.1850281786 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000030_200 |
0.1854728051 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 15 |
Hb_007313_080 |
0.1867334584 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 16 |
Hb_001059_120 |
0.1870207024 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_147752_010 |
0.1901908776 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 18 |
Hb_002391_220 |
0.1906893967 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 19 |
Hb_000445_490 |
0.1907754592 |
- |
- |
- |
| 20 |
Hb_002774_050 |
0.1924907628 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |