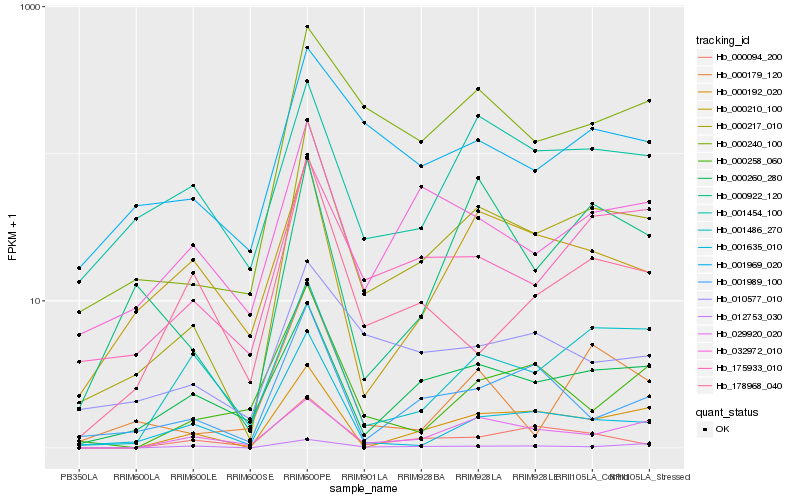
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000217_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 2 |
Hb_000240_100 |
0.1740942511 |
- |
- |
PREDICTED: ras-related protein Rab5-like [Eucalyptus grandis] |
| 3 |
Hb_000260_280 |
0.1767078574 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 4 |
Hb_000192_020 |
0.179908578 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 5 |
Hb_175933_010 |
0.1815544853 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 6 |
Hb_029920_020 |
0.2079969925 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 7 |
Hb_032972_010 |
0.2117727032 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 8 |
Hb_001989_100 |
0.2145240709 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 9 |
Hb_001969_020 |
0.2161887908 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 10 |
Hb_001454_100 |
0.2184567295 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 11 |
Hb_000179_120 |
0.2193759352 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 12 |
Hb_001486_270 |
0.2194147117 |
- |
- |
Ann10 [Manihot esculenta] |
| 13 |
Hb_000922_120 |
0.2213667024 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 14 |
Hb_001635_010 |
0.2228923994 |
- |
- |
- |
| 15 |
Hb_010577_010 |
0.2295072613 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 16 |
Hb_000210_100 |
0.230288017 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 17 |
Hb_012753_030 |
0.2320805811 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 18 |
Hb_000094_200 |
0.2326873166 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000258_060 |
0.2337442857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_178968_040 |
0.2365464576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |