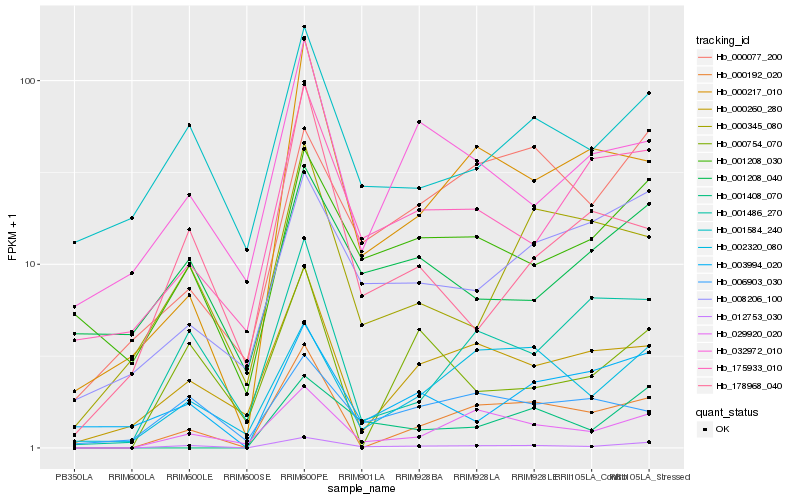
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012753_030 |
0.0 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 2 |
Hb_029920_020 |
0.1817552061 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 3 |
Hb_001208_030 |
0.2031800577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000192_020 |
0.2035366036 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 5 |
Hb_001584_240 |
0.2107430137 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 6 |
Hb_001486_270 |
0.2179879646 |
- |
- |
Ann10 [Manihot esculenta] |
| 7 |
Hb_175933_010 |
0.2220147784 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 8 |
Hb_000260_280 |
0.2225165285 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 9 |
Hb_002320_080 |
0.2262043288 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003994_020 |
0.2299347917 |
- |
- |
- |
| 11 |
Hb_000217_010 |
0.2320805811 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 12 |
Hb_008206_100 |
0.2323395601 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 13 |
Hb_000754_070 |
0.2341536962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001208_040 |
0.2361270671 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 15 |
Hb_032972_010 |
0.2378501876 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 16 |
Hb_000345_080 |
0.241060503 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 17 |
Hb_001408_070 |
0.2426905706 |
- |
- |
- |
| 18 |
Hb_006903_030 |
0.2461160952 |
- |
- |
PREDICTED: uncharacterized protein LOC105640666 [Jatropha curcas] |
| 19 |
Hb_000077_200 |
0.2486505535 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 20 |
Hb_178968_040 |
0.2497503633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |