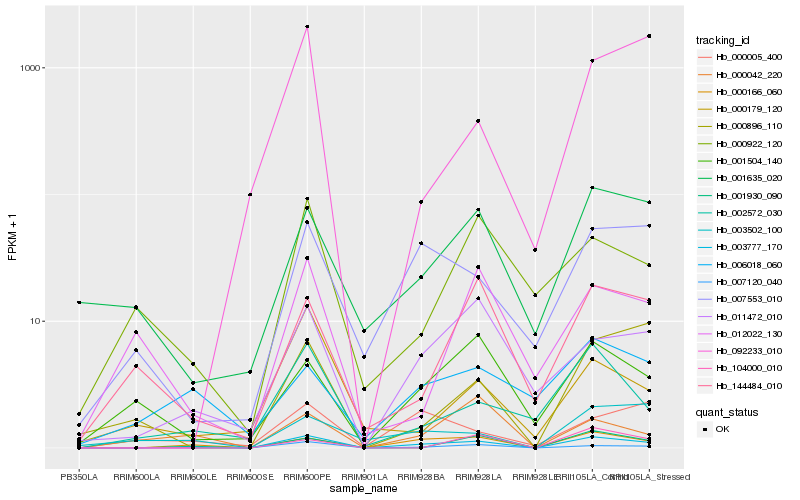
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003777_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000166_060 |
0.1653479031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007120_040 |
0.1956483182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011472_010 |
0.2511749723 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 5 |
Hb_001930_090 |
0.2600568768 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_002572_030 |
0.2628824088 |
- |
- |
PREDICTED: diacylglycerol kinase theta [Jatropha curcas] |
| 7 |
Hb_007553_010 |
0.2639993991 |
- |
- |
- |
| 8 |
Hb_001635_020 |
0.2751586408 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 9 |
Hb_001504_140 |
0.2819442593 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 10 |
Hb_144484_010 |
0.2852081496 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_000042_220 |
0.2945115755 |
- |
- |
PREDICTED: uncharacterized protein LOC105632809 [Jatropha curcas] |
| 12 |
Hb_000922_120 |
0.2974718671 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 13 |
Hb_003502_100 |
0.2980320681 |
- |
- |
- |
| 14 |
Hb_000005_400 |
0.2988732298 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 23 isoform X2 [Jatropha curcas] |
| 15 |
Hb_092233_010 |
0.3022445116 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 16 |
Hb_006018_060 |
0.3039347708 |
- |
- |
Zinc finger SWIM domain-containing protein 7 isoform 1 [Theobroma cacao] |
| 17 |
Hb_104000_010 |
0.3053662888 |
- |
- |
- |
| 18 |
Hb_000896_110 |
0.3056617979 |
- |
- |
- |
| 19 |
Hb_000179_120 |
0.3099880509 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 20 |
Hb_012022_130 |
0.3129477838 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |