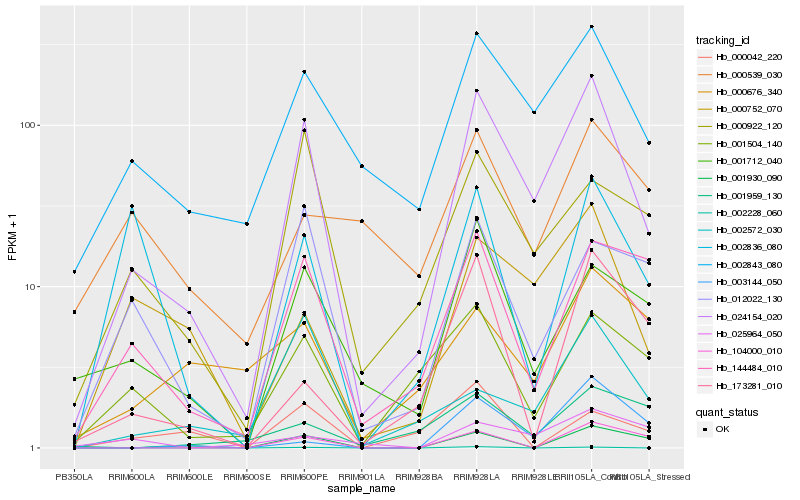
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024154_020 |
0.0 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 2 |
Hb_002843_080 |
0.2026988685 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 3 |
Hb_000752_070 |
0.2443412442 |
- |
- |
PREDICTED: mavicyanin-like [Jatropha curcas] |
| 4 |
Hb_001712_040 |
0.2503446919 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 5 |
Hb_144484_010 |
0.2536627243 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 6 |
Hb_000922_120 |
0.255220465 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 7 |
Hb_001504_140 |
0.2613418065 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 8 |
Hb_012022_130 |
0.2630790053 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 9 |
Hb_001930_090 |
0.2725557315 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_002572_030 |
0.2868701314 |
- |
- |
PREDICTED: diacylglycerol kinase theta [Jatropha curcas] |
| 11 |
Hb_002228_060 |
0.2922187487 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 12 |
Hb_104000_010 |
0.2926160962 |
- |
- |
- |
| 13 |
Hb_002836_080 |
0.2965633652 |
- |
- |
- |
| 14 |
Hb_173281_010 |
0.3016614983 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000676_340 |
0.3030716308 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_003144_050 |
0.3040761524 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 17 |
Hb_025964_050 |
0.3070323565 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 18 |
Hb_001959_130 |
0.3104457384 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_000539_030 |
0.3143894703 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 20 |
Hb_000042_220 |
0.3208133325 |
- |
- |
PREDICTED: uncharacterized protein LOC105632809 [Jatropha curcas] |