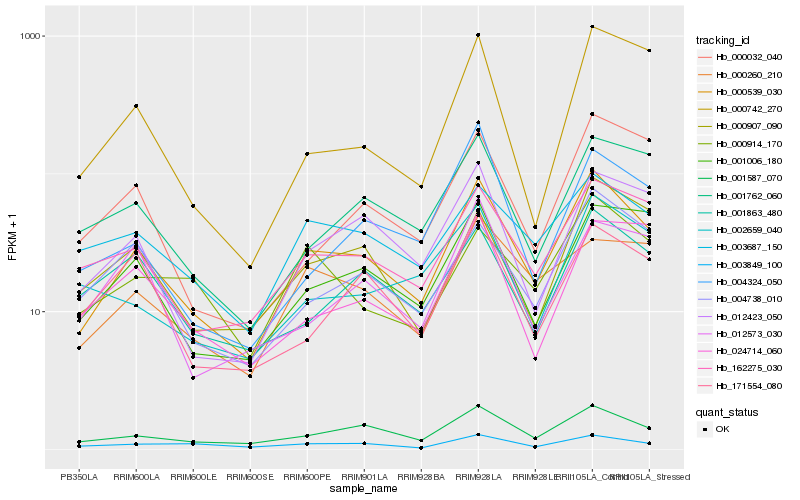
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000539_030 |
0.0 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 2 |
Hb_001587_070 |
0.1127229279 |
desease resistance |
Gene Name: NB-ARC |
Nbs-lrr resistance protein, putative [Theobroma cacao] |
| 3 |
Hb_162275_030 |
0.12969389 |
- |
- |
PREDICTED: uncharacterized protein LOC105646638 [Jatropha curcas] |
| 4 |
Hb_012423_050 |
0.1383927652 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 5 |
Hb_001006_180 |
0.1395912641 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |
| 6 |
Hb_004738_010 |
0.1435193065 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 7 |
Hb_000907_090 |
0.147188629 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_001762_060 |
0.1499856407 |
- |
- |
pantothenate kinase, putative [Ricinus communis] |
| 9 |
Hb_003687_150 |
0.1543040909 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000260_210 |
0.1556005365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000032_040 |
0.156001191 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 12 |
Hb_001863_480 |
0.160656643 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 13 |
Hb_171554_080 |
0.1630341845 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 14 |
Hb_000914_170 |
0.1642215721 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 15 |
Hb_000742_270 |
0.1659705813 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 16 |
Hb_002659_040 |
0.1675124877 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_004324_050 |
0.1682864742 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 18 |
Hb_003849_100 |
0.1696741215 |
- |
- |
PREDICTED: putative transcription elongation factor SPT5 homolog 1 [Populus euphratica] |
| 19 |
Hb_024714_060 |
0.1703961141 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012573_030 |
0.172156583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |