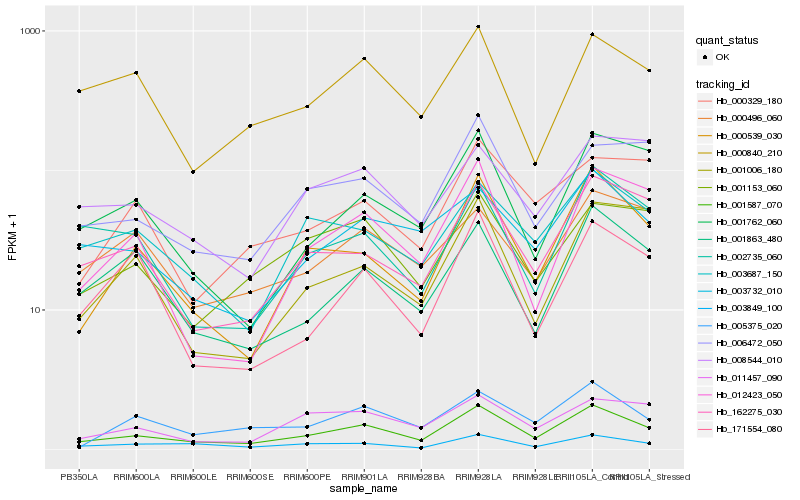
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001587_070 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Nbs-lrr resistance protein, putative [Theobroma cacao] |
| 2 |
Hb_000539_030 |
0.1127229279 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 3 |
Hb_162275_030 |
0.1278947025 |
- |
- |
PREDICTED: uncharacterized protein LOC105646638 [Jatropha curcas] |
| 4 |
Hb_003849_100 |
0.1303387463 |
- |
- |
PREDICTED: putative transcription elongation factor SPT5 homolog 1 [Populus euphratica] |
| 5 |
Hb_006472_050 |
0.1328385709 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 6 |
Hb_003732_010 |
0.1331839082 |
- |
- |
hypothetical protein CHLNCDRAFT_58243 [Chlorella variabilis] |
| 7 |
Hb_003687_150 |
0.1334177047 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001153_060 |
0.1342007303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001863_480 |
0.1399523979 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 10 |
Hb_012423_050 |
0.1399804684 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 11 |
Hb_001762_060 |
0.1403414186 |
- |
- |
pantothenate kinase, putative [Ricinus communis] |
| 12 |
Hb_011457_090 |
0.1413826306 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 13 |
Hb_005375_020 |
0.1414900577 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 1-like [Nicotiana sylvestris] |
| 14 |
Hb_000840_210 |
0.1449408979 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 15 |
Hb_008544_010 |
0.1476192997 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000496_060 |
0.1487833653 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |
| 17 |
Hb_000329_180 |
0.1515967895 |
- |
- |
Auxin-induced protein X10A, putative [Ricinus communis] |
| 18 |
Hb_171554_080 |
0.1520890419 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 19 |
Hb_002735_060 |
0.1525869173 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 20 |
Hb_001006_180 |
0.1539781989 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |