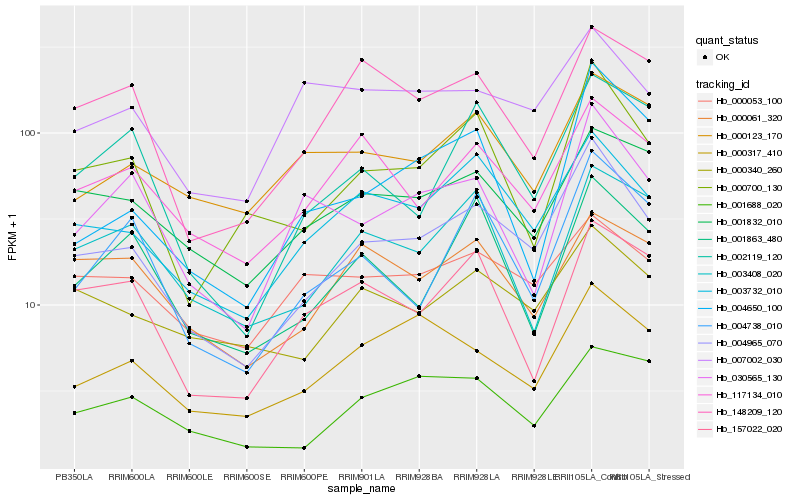
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004965_070 |
0.0 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 2 |
Hb_003732_010 |
0.1064351405 |
- |
- |
hypothetical protein CHLNCDRAFT_58243 [Chlorella variabilis] |
| 3 |
Hb_000700_130 |
0.1300435077 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000340_260 |
0.1355527146 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 5 |
Hb_001832_010 |
0.1449737727 |
- |
- |
PREDICTED: uncharacterized protein LOC105637890 isoform X1 [Jatropha curcas] |
| 6 |
Hb_117134_010 |
0.1479368002 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_004738_010 |
0.1485144693 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 8 |
Hb_000317_410 |
0.1489712789 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 9 |
Hb_148209_120 |
0.1520849193 |
- |
- |
hypothetical protein CISIN_1g032846mg [Citrus sinensis] |
| 10 |
Hb_002119_120 |
0.1531387427 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_001688_020 |
0.1546309582 |
- |
- |
PREDICTED: uncharacterized protein LOC103438026 [Malus domestica] |
| 12 |
Hb_003408_020 |
0.1591980089 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 13 |
Hb_000053_100 |
0.1595218988 |
- |
- |
PREDICTED: uncharacterized protein LOC105642071 [Jatropha curcas] |
| 14 |
Hb_001863_480 |
0.1600434844 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 15 |
Hb_004650_100 |
0.1600941583 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_030565_130 |
0.1610524496 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 17 |
Hb_000061_320 |
0.1612250038 |
- |
- |
PREDICTED: uncharacterized protein LOC105640575 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007002_030 |
0.1616767583 |
- |
- |
PREDICTED: transmembrane protein 205 [Jatropha curcas] |
| 19 |
Hb_000123_170 |
0.162213405 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 20 |
Hb_157022_020 |
0.1630301891 |
- |
- |
nicotinamide mononucleotide adenylyltransferase, putative [Ricinus communis] |