| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
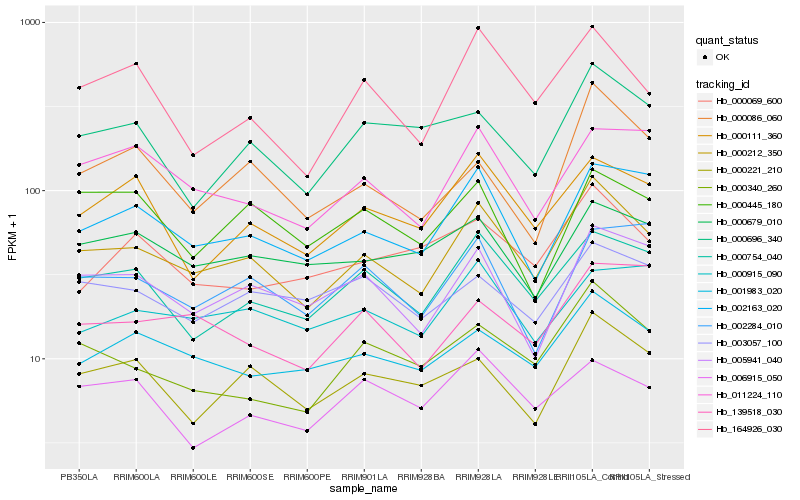
Hb_000212_350 |
0.0 |
- |
- |
glucosamine-fructose-6-phosphate aminotransferase, putative [Ricinus communis] |
| 2 |
Hb_000754_040 |
0.0950011309 |
- |
- |
PREDICTED: uncharacterized protein LOC105115746 [Populus euphratica] |
| 3 |
Hb_002163_020 |
0.0966727303 |
transcription factor |
TF Family: PHD |
PREDICTED: BAH and coiled-coil domain-containing protein 1-like [Jatropha curcas] |
| 4 |
Hb_000340_260 |
0.1033790573 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 5 |
Hb_001983_020 |
0.1035103093 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 6 |
Hb_005941_040 |
0.1041311924 |
- |
- |
PREDICTED: CASP-like protein 5A2 [Jatropha curcas] |
| 7 |
Hb_164926_030 |
0.1041486805 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 8 |
Hb_000221_210 |
0.1092329208 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 9 |
Hb_000111_360 |
0.1112978526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011224_110 |
0.1140898165 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 11 |
Hb_000696_340 |
0.1192557735 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 12 |
Hb_000679_010 |
0.1202152117 |
- |
- |
PREDICTED: uncharacterized protein LOC105637663 [Jatropha curcas] |
| 13 |
Hb_000086_060 |
0.121651385 |
- |
- |
PREDICTED: TOM1-like protein 2 isoform X2 [Populus euphratica] |
| 14 |
Hb_000069_600 |
0.1241487946 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 15 |
Hb_003057_100 |
0.1243562289 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: S-adenosylmethionine mitochondrial carrier protein-like [Malus domestica] |
| 16 |
Hb_000915_090 |
0.1254343582 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b [Jatropha curcas] |
| 17 |
Hb_139518_030 |
0.1276196223 |
- |
- |
protein kinase atn1, putative [Ricinus communis] |
| 18 |
Hb_006915_050 |
0.1276259182 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002284_010 |
0.1284738249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000445_180 |
0.1310480493 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |