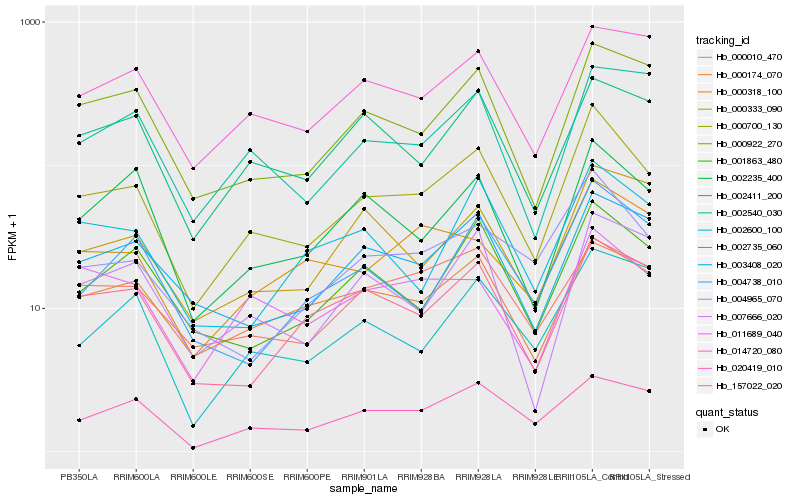
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000922_270 |
0.1238168092 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 3 |
Hb_000174_070 |
0.1259893272 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_003408_020 |
0.1292422187 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 5 |
Hb_001863_480 |
0.1294831371 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 6 |
Hb_004965_070 |
0.1300435077 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 7 |
Hb_002235_400 |
0.1321209324 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 8 |
Hb_020419_010 |
0.1332427754 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 9 |
Hb_157022_020 |
0.1351703332 |
- |
- |
nicotinamide mononucleotide adenylyltransferase, putative [Ricinus communis] |
| 10 |
Hb_002411_200 |
0.1389397191 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_014720_080 |
0.1393973577 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 12 |
Hb_007666_020 |
0.1407154957 |
- |
- |
alkaline phytoceramidase, putative [Ricinus communis] |
| 13 |
Hb_002600_100 |
0.1438090014 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 14 |
Hb_011689_040 |
0.1457867085 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 15 |
Hb_002735_060 |
0.1461125633 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 16 |
Hb_000318_100 |
0.1465380692 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 17 |
Hb_000333_090 |
0.1473874496 |
- |
- |
PREDICTED: uncharacterized protein LOC102617374 isoform X1 [Citrus sinensis] |
| 18 |
Hb_000010_470 |
0.1474433747 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_004738_010 |
0.1475116306 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 20 |
Hb_002540_030 |
0.148498587 |
- |
- |
hypothetical protein 32 [Hevea brasiliensis] |