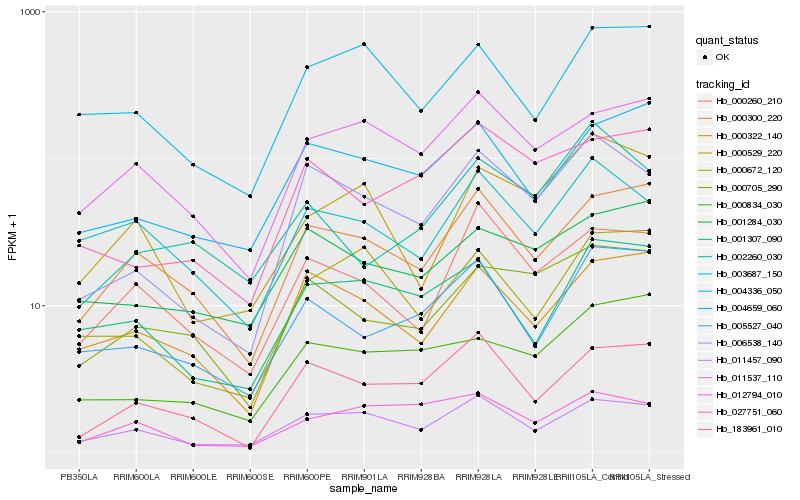
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103341483 [Prunus mume] |
| 2 |
Hb_011457_090 |
0.1334349083 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 3 |
Hb_183961_010 |
0.1438023366 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Cucumis sativus] |
| 4 |
Hb_027751_060 |
0.1493639645 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 5 |
Hb_000322_140 |
0.151582166 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 6 |
Hb_000705_290 |
0.1547874239 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 7 |
Hb_000260_210 |
0.1637830549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000300_220 |
0.164051196 |
- |
- |
PREDICTED: uncharacterized protein LOC105632569 [Jatropha curcas] |
| 9 |
Hb_001284_030 |
0.1695137228 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000672_120 |
0.1697855309 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 11 |
Hb_001307_090 |
0.1702444412 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 12 |
Hb_004659_060 |
0.1716409277 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 13 |
Hb_011537_110 |
0.1721711493 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 14 |
Hb_005527_040 |
0.1723227726 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 15 |
Hb_003687_150 |
0.1748129319 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000529_220 |
0.17504866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000834_030 |
0.1773919367 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004336_050 |
0.1789688234 |
- |
- |
hypothetical protein JCGZ_26651 [Jatropha curcas] |
| 19 |
Hb_012794_010 |
0.1793169292 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 20 |
Hb_002260_030 |
0.1794006714 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |