| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
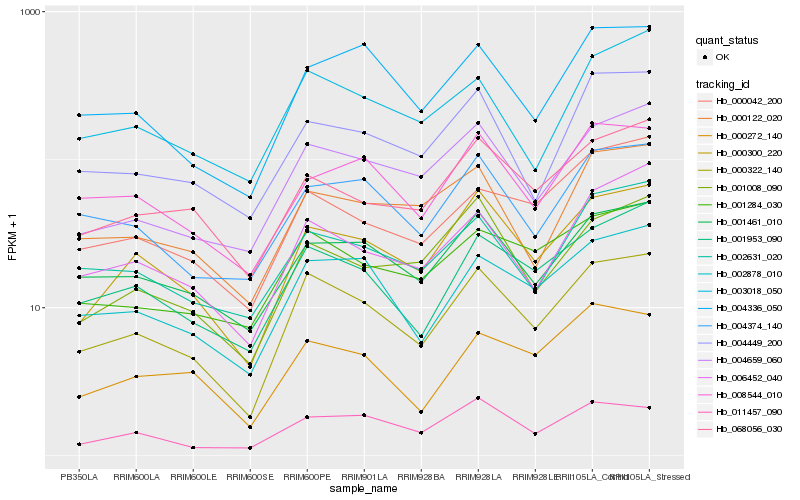
Hb_000322_140 |
0.0 |
- |
- |
hypothetical protein RCOM_1047200 [Ricinus communis] |
| 2 |
Hb_000300_220 |
0.0795115119 |
- |
- |
PREDICTED: uncharacterized protein LOC105632569 [Jatropha curcas] |
| 3 |
Hb_002878_010 |
0.0868512447 |
- |
- |
- |
| 4 |
Hb_001953_090 |
0.0920948498 |
- |
- |
PREDICTED: membrane-anchored ubiquitin-fold protein 3 [Jatropha curcas] |
| 5 |
Hb_004659_060 |
0.096197221 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 6 |
Hb_001461_010 |
0.0981489085 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Pyrus x bretschneideri] |
| 7 |
Hb_004374_140 |
0.1042373822 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 8 |
Hb_001284_030 |
0.10533248 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_004336_050 |
0.1059328379 |
- |
- |
hypothetical protein JCGZ_26651 [Jatropha curcas] |
| 10 |
Hb_006452_040 |
0.1061383631 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 11 |
Hb_002631_020 |
0.1078696096 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 12 |
Hb_011457_090 |
0.1103853743 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 13 |
Hb_068056_030 |
0.1105277619 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 14 |
Hb_004449_200 |
0.1118631767 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 1 [Populus euphratica] |
| 15 |
Hb_001008_090 |
0.1123181608 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 16 |
Hb_003018_050 |
0.1127581201 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase Pin1 [Jatropha curcas] |
| 17 |
Hb_000042_200 |
0.1138025586 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |
| 18 |
Hb_000272_140 |
0.11394725 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 19 |
Hb_008544_010 |
0.1143823709 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000122_020 |
0.1144600198 |
- |
- |
PREDICTED: alpha-soluble NSF attachment protein 2 [Jatropha curcas] |