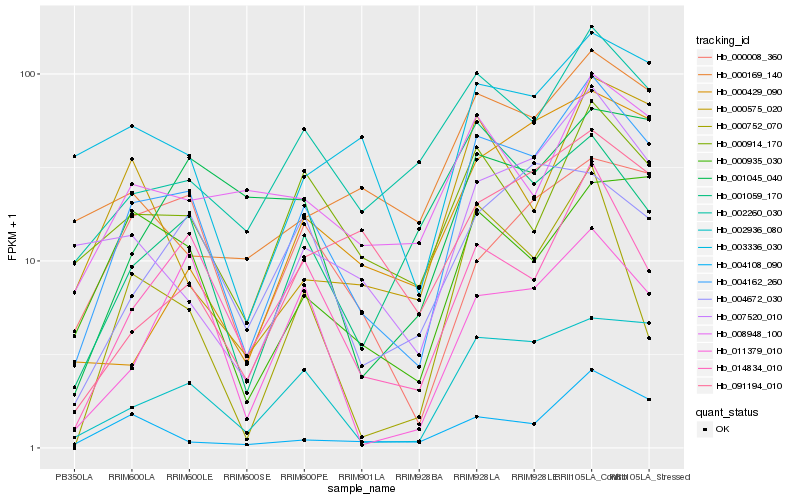
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_260 |
0.0 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002936_080 |
0.1482675411 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 3 |
Hb_014834_010 |
0.1602806038 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4 [Jatropha curcas] |
| 4 |
Hb_002260_030 |
0.1815207035 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 5 |
Hb_007520_010 |
0.1845062555 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 6 |
Hb_000914_170 |
0.1878472094 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 7 |
Hb_000935_030 |
0.1987320428 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_003336_030 |
0.1995271666 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_004108_090 |
0.2001725889 |
- |
- |
PREDICTED: uncharacterized protein LOC105650231 [Jatropha curcas] |
| 10 |
Hb_008948_100 |
0.2044327934 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 11 |
Hb_000752_070 |
0.2092368402 |
- |
- |
PREDICTED: mavicyanin-like [Jatropha curcas] |
| 12 |
Hb_000169_140 |
0.2115060272 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 13 |
Hb_000008_360 |
0.2117536662 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 14 |
Hb_011379_010 |
0.211960037 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000575_020 |
0.2133562363 |
- |
- |
PREDICTED: copper-transporting ATPase PAA1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004672_030 |
0.2147444496 |
- |
- |
proline oxidase [Manihot esculenta] |
| 17 |
Hb_091194_010 |
0.2156480598 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 18 |
Hb_001045_040 |
0.2170586966 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 2 [Jatropha curcas] |
| 19 |
Hb_001059_170 |
0.2197793294 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 20 |
Hb_000429_090 |
0.2220754622 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 3, chloroplastic [Jatropha curcas] |