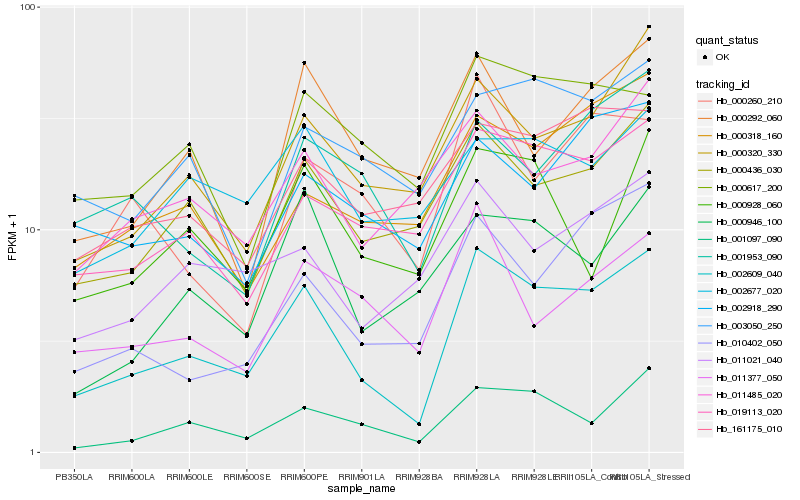
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_040 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 2 |
Hb_001097_090 |
0.1480984132 |
- |
- |
Translation initiation factor SUI1 family protein, putative [Theobroma cacao] |
| 3 |
Hb_000436_030 |
0.148605298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_019113_020 |
0.1507165985 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 5 |
Hb_000320_330 |
0.1541587886 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 6 |
Hb_010402_050 |
0.1615454442 |
- |
- |
- |
| 7 |
Hb_011021_040 |
0.1655770926 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 8 |
Hb_161175_010 |
0.1685736563 |
- |
- |
PREDICTED: vesicle-associated protein 1-1 [Jatropha curcas] |
| 9 |
Hb_000318_160 |
0.1692652585 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |
| 10 |
Hb_000617_200 |
0.1702277021 |
- |
- |
f-actin capping protein alpha, putative [Ricinus communis] |
| 11 |
Hb_000946_100 |
0.1716247034 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 12 |
Hb_000292_060 |
0.1722807274 |
- |
- |
transporter, putative [Ricinus communis] |
| 13 |
Hb_011485_020 |
0.1732573223 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_003050_250 |
0.1737511032 |
- |
- |
PREDICTED: uncharacterized protein At2g34460, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002677_020 |
0.1741486593 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 16 |
Hb_000928_060 |
0.1747558139 |
- |
- |
transporter, putative [Ricinus communis] |
| 17 |
Hb_000260_210 |
0.1747661218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011377_050 |
0.1747681602 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001953_090 |
0.1749208508 |
- |
- |
PREDICTED: membrane-anchored ubiquitin-fold protein 3 [Jatropha curcas] |
| 20 |
Hb_002918_290 |
0.1753325458 |
- |
- |
PREDICTED: uncharacterized protein LOC105649583 isoform X1 [Jatropha curcas] |