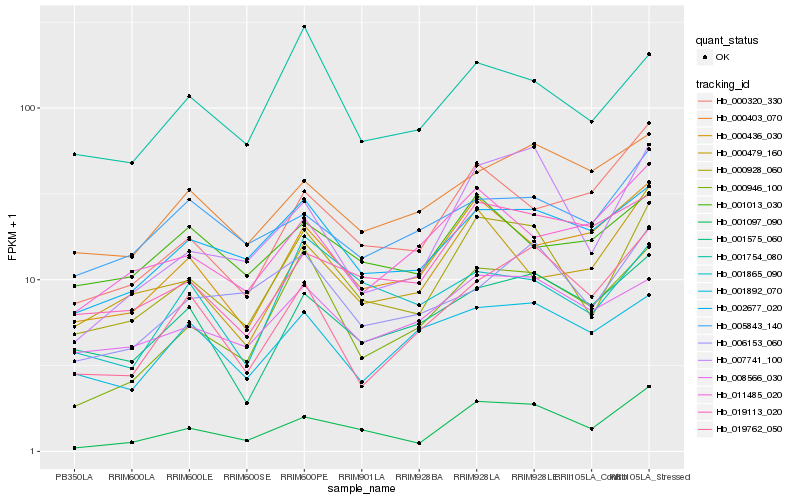
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_060 |
0.0 |
- |
- |
transporter, putative [Ricinus communis] |
| 2 |
Hb_001097_090 |
0.120545921 |
- |
- |
Translation initiation factor SUI1 family protein, putative [Theobroma cacao] |
| 3 |
Hb_007741_100 |
0.124385565 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 4 |
Hb_000946_100 |
0.1309363637 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_002677_020 |
0.1316502434 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 6 |
Hb_001754_080 |
0.1349091318 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 7 |
Hb_008566_030 |
0.1361435584 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 8 |
Hb_000436_030 |
0.1445261515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001575_060 |
0.1454200055 |
- |
- |
PREDICTED: 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_006153_060 |
0.1493769458 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_000479_160 |
0.150888077 |
- |
- |
Membrane-anchored ubiquitin-fold protein 1 precursor isoform 2 [Theobroma cacao] |
| 12 |
Hb_001892_070 |
0.1566692439 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 13 |
Hb_001865_090 |
0.1579330361 |
- |
- |
hypothetical protein VITISV_005773 [Vitis vinifera] |
| 14 |
Hb_019113_020 |
0.158639378 |
- |
- |
hypothetical protein JCGZ_13398 [Jatropha curcas] |
| 15 |
Hb_000403_070 |
0.1596385768 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_011485_020 |
0.1598360046 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001013_030 |
0.1603967962 |
- |
- |
GTP-binding protein yptv3, putative [Ricinus communis] |
| 18 |
Hb_019762_050 |
0.1607731002 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 19 |
Hb_005843_140 |
0.162211458 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000320_330 |
0.1642248959 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |