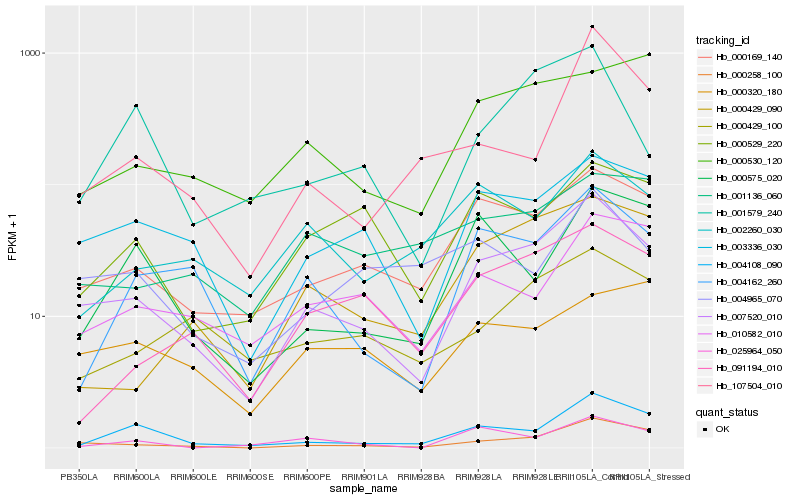
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007520_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 2 |
Hb_000258_100 |
0.1342140898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000169_140 |
0.149121753 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 4 |
Hb_003336_030 |
0.1561266227 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_004108_090 |
0.1843996529 |
- |
- |
PREDICTED: uncharacterized protein LOC105650231 [Jatropha curcas] |
| 6 |
Hb_004162_260 |
0.1845062555 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010582_010 |
0.2041673303 |
- |
- |
hypothetical protein POPTR_0011s15670g [Populus trichocarpa] |
| 8 |
Hb_000429_090 |
0.205476729 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_091194_010 |
0.2059798473 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 10 |
Hb_000429_100 |
0.2077546818 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 11 |
Hb_000530_120 |
0.2095250147 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 12 |
Hb_000529_220 |
0.2107707387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002260_030 |
0.2109881189 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 14 |
Hb_001579_240 |
0.2138041524 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 15 |
Hb_000575_020 |
0.2195727073 |
- |
- |
PREDICTED: copper-transporting ATPase PAA1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000320_180 |
0.2202087923 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 17 |
Hb_025964_050 |
0.2209781436 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 18 |
Hb_107504_010 |
0.2223562705 |
- |
- |
- |
| 19 |
Hb_001136_060 |
0.2224194013 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 20 |
Hb_004965_070 |
0.2237797404 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |