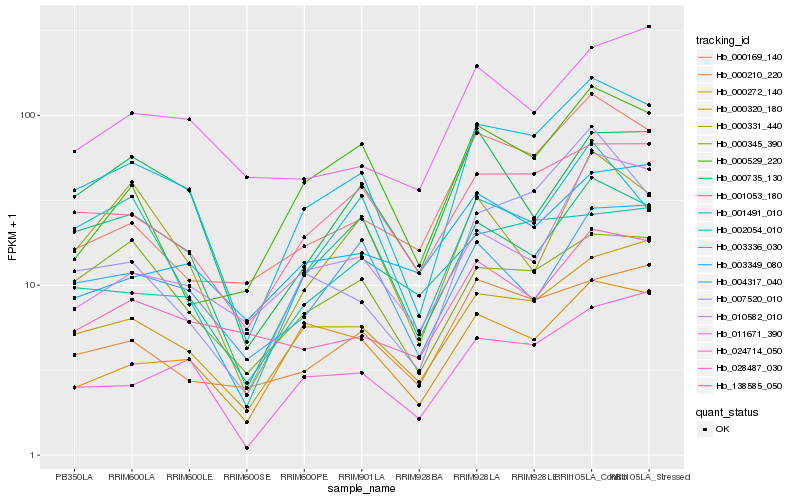
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003336_030 |
0.0 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_138585_050 |
0.1202623069 |
- |
- |
ubiquitin-conjugating enzyme E2-25kD, putative [Ricinus communis] |
| 3 |
Hb_000320_180 |
0.1224238788 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 4 |
Hb_024714_050 |
0.12745101 |
- |
- |
hypothetical protein CICLE_v100188992mg, partial [Citrus clementina] |
| 5 |
Hb_000210_220 |
0.1472481945 |
- |
- |
hypothetical protein POPTR_0006s22010g [Populus trichocarpa] |
| 6 |
Hb_000272_140 |
0.1492285669 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 7 |
Hb_000169_140 |
0.1495322157 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 8 |
Hb_028487_030 |
0.1532130624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010582_010 |
0.154636787 |
- |
- |
hypothetical protein POPTR_0011s15670g [Populus trichocarpa] |
| 10 |
Hb_001053_180 |
0.1551780954 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_004317_040 |
0.1560835877 |
- |
- |
hypothetical protein POPTR_0006s22010g [Populus trichocarpa] |
| 12 |
Hb_007520_010 |
0.1561266227 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 13 |
Hb_002054_010 |
0.1563249706 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 14 |
Hb_000345_390 |
0.1566738651 |
- |
- |
hypothetical protein JCGZ_23349 [Jatropha curcas] |
| 15 |
Hb_001491_010 |
0.1575516249 |
- |
- |
PREDICTED: uncharacterized protein LOC105649805 [Jatropha curcas] |
| 16 |
Hb_000331_440 |
0.1635469712 |
- |
- |
hypothetical protein POPTR_0015s10290g [Populus trichocarpa] |
| 17 |
Hb_003349_080 |
0.1649123688 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 18 |
Hb_000529_220 |
0.1650523886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000735_130 |
0.1652655025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011671_390 |
0.1658165077 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |