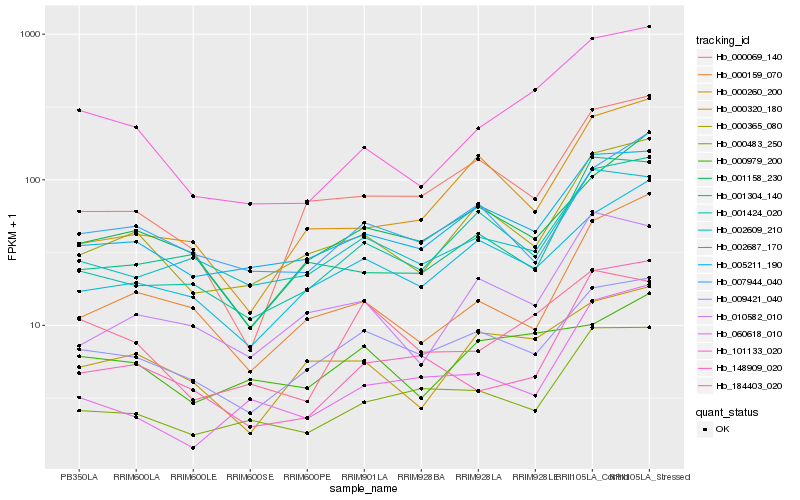
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101133_020 |
0.0 |
- |
- |
PREDICTED: 15.7 kDa heat shock protein, peroxisomal [Jatropha curcas] |
| 2 |
Hb_000365_080 |
0.1529111335 |
- |
- |
PREDICTED: uncharacterized protein LOC105649037 [Jatropha curcas] |
| 3 |
Hb_005211_190 |
0.1574844577 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 4 |
Hb_001424_020 |
0.1680639286 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 isoform X2 [Solanum lycopersicum] |
| 5 |
Hb_000320_180 |
0.1681596983 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 6 |
Hb_148909_020 |
0.1697137848 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_000159_070 |
0.174325815 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 8 |
Hb_000483_250 |
0.17607567 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 9 |
Hb_000069_140 |
0.1801179394 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 10 |
Hb_001304_140 |
0.180780809 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 11 |
Hb_009421_040 |
0.1822965665 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 12 |
Hb_001158_230 |
0.1830324125 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 13 |
Hb_184403_020 |
0.1835236921 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Vitis vinifera] |
| 14 |
Hb_010582_010 |
0.1843873637 |
- |
- |
hypothetical protein POPTR_0011s15670g [Populus trichocarpa] |
| 15 |
Hb_002687_170 |
0.186020232 |
- |
- |
PREDICTED: G patch domain and ankyrin repeat-containing protein 1 homolog [Jatropha curcas] |
| 16 |
Hb_060618_010 |
0.1883057373 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 17 |
Hb_000260_200 |
0.1884121368 |
- |
- |
- |
| 18 |
Hb_002609_210 |
0.1886445594 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_000979_200 |
0.1890071404 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 20 |
Hb_007944_040 |
0.1898036226 |
- |
- |
60S ribosomal protein L18aB [Hevea brasiliensis] |