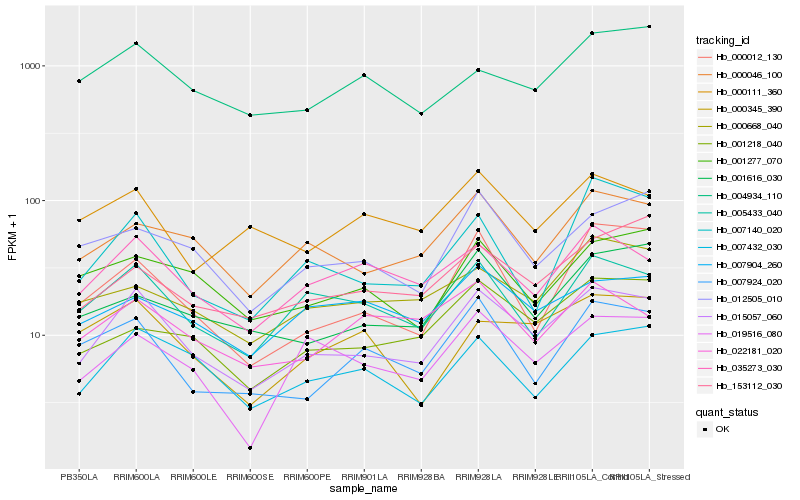
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015057_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_005433_040 |
0.1135322957 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 3 |
Hb_000046_100 |
0.1186624633 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_035273_030 |
0.1213413917 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 5 |
Hb_007432_030 |
0.1223677954 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 6 |
Hb_019516_080 |
0.1339278198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007924_020 |
0.136537036 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_001218_040 |
0.1367056422 |
- |
- |
Mitochondrial substrate carrier family protein [Theobroma cacao] |
| 9 |
Hb_000012_130 |
0.1376948477 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 10 |
Hb_012505_010 |
0.1391813054 |
- |
- |
mak, putative [Ricinus communis] |
| 11 |
Hb_001277_070 |
0.1402289405 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_153112_030 |
0.1405376305 |
- |
- |
PREDICTED: UPF0678 fatty acid-binding protein-like protein At1g79260 [Jatropha curcas] |
| 13 |
Hb_007904_260 |
0.143466391 |
- |
- |
PREDICTED: uncharacterized protein LOC105644715 [Jatropha curcas] |
| 14 |
Hb_000668_040 |
0.1440452796 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 15 |
Hb_022181_020 |
0.1450753095 |
- |
- |
PREDICTED: D-tyrosyl-tRNA(Tyr) deacylase isoform X1 [Jatropha curcas] |
| 16 |
Hb_000345_390 |
0.1453645222 |
- |
- |
hypothetical protein JCGZ_23349 [Jatropha curcas] |
| 17 |
Hb_004934_110 |
0.1455652875 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 18 |
Hb_001616_030 |
0.1464078882 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor-like protein At4g13040 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007140_020 |
0.1465114341 |
- |
- |
hypothetical protein CICLE_v10008419mg [Citrus clementina] |
| 20 |
Hb_000111_360 |
0.1467307439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |