| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
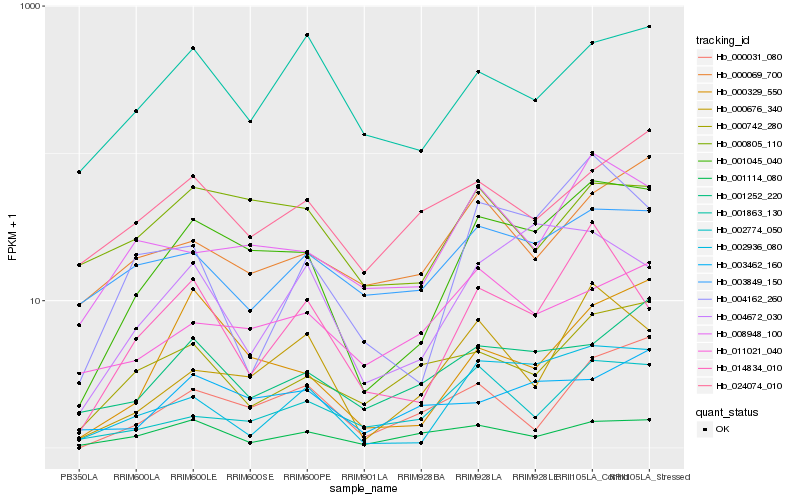
Hb_001045_040 |
0.0 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 2 [Jatropha curcas] |
| 2 |
Hb_002936_080 |
0.1831562389 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 3 |
Hb_000329_550 |
0.1841583569 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 4 |
Hb_008948_100 |
0.1917866838 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_000676_340 |
0.2015237673 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_001252_220 |
0.204503035 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 7 |
Hb_003462_160 |
0.2051203033 |
transcription factor |
TF Family: GNAT |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 8 |
Hb_004672_030 |
0.2106218279 |
- |
- |
proline oxidase [Manihot esculenta] |
| 9 |
Hb_000031_080 |
0.211192257 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 10 |
Hb_002774_050 |
0.2115164686 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 11 |
Hb_000742_280 |
0.2123516298 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_014834_010 |
0.2127647583 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4 [Jatropha curcas] |
| 13 |
Hb_000805_110 |
0.2131848198 |
- |
- |
PREDICTED: uncharacterized protein C24B11.05 [Jatropha curcas] |
| 14 |
Hb_011021_040 |
0.2140497104 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 15 |
Hb_001863_130 |
0.2158171528 |
- |
- |
hypothetical protein ZEAMMB73_433257, partial [Zea mays] |
| 16 |
Hb_003849_150 |
0.2170095868 |
- |
- |
PREDICTED: uncharacterized protein LOC105644698 [Jatropha curcas] |
| 17 |
Hb_004162_260 |
0.2170586966 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000069_700 |
0.217994718 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 19 |
Hb_001114_080 |
0.2181535171 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 20 |
Hb_024074_010 |
0.2212281892 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |