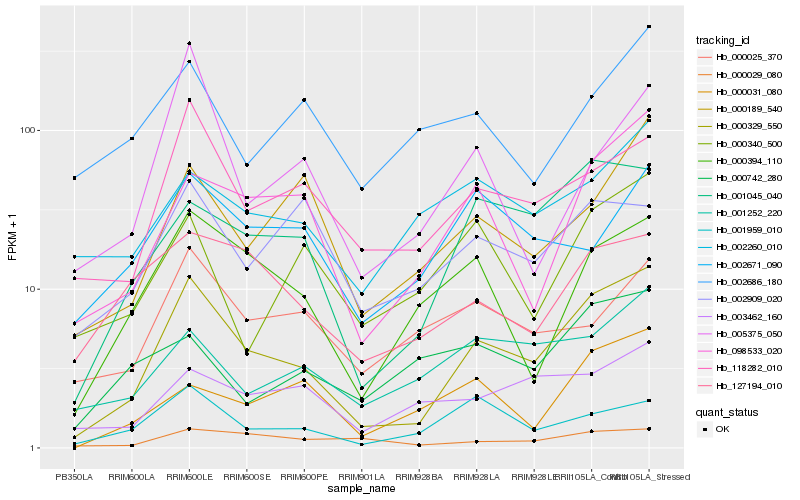
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_550 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 8 [Jatropha curcas] |
| 2 |
Hb_001045_040 |
0.1841583569 |
- |
- |
PREDICTED: carbonyl reductase [NADPH] 2 [Jatropha curcas] |
| 3 |
Hb_098533_020 |
0.1887476177 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 4 |
Hb_118282_010 |
0.198839494 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 5 |
Hb_000394_110 |
0.2017196024 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000189_540 |
0.2135683188 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001959_010 |
0.2174264079 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 8 |
Hb_001252_220 |
0.2218773178 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 9 |
Hb_127194_010 |
0.2270456081 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 10 |
Hb_003462_160 |
0.2291435063 |
transcription factor |
TF Family: GNAT |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_000031_080 |
0.2292707739 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 12 |
Hb_000340_500 |
0.2299304847 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 13 |
Hb_002260_010 |
0.2313457051 |
- |
- |
PREDICTED: coiled-coil-helix-coiled-coil-helix domain-containing protein 10, mitochondrial [Sesamum indicum] |
| 14 |
Hb_000025_370 |
0.2351197703 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000029_080 |
0.2377512562 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 16 |
Hb_005375_050 |
0.2385330523 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 17 |
Hb_002671_090 |
0.2387472441 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 18 |
Hb_002909_020 |
0.2434852202 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 19 |
Hb_002686_180 |
0.2438711292 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 20 |
Hb_000742_280 |
0.2449511082 |
- |
- |
ATP binding protein, putative [Ricinus communis] |