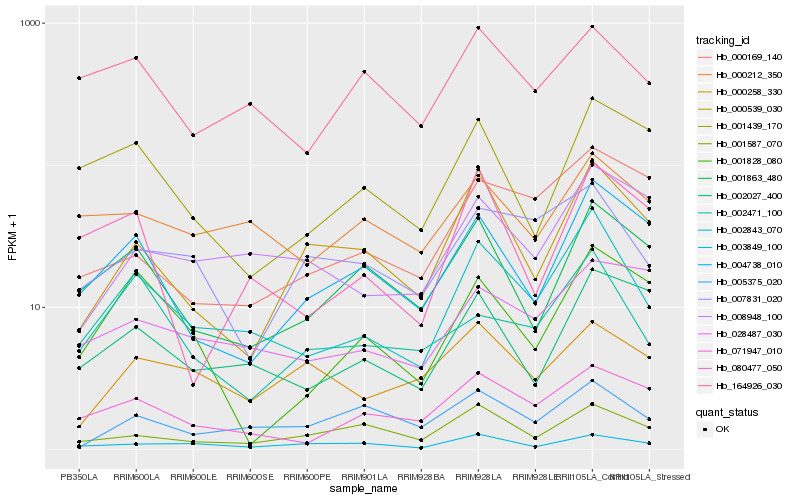
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002843_070 |
0.0 |
- |
- |
unknown [Lotus japonicus] |
| 2 |
Hb_071947_010 |
0.1798864801 |
- |
- |
- |
| 3 |
Hb_008948_100 |
0.1837813841 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 4 |
Hb_004738_010 |
0.1874486754 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 5 |
Hb_002027_400 |
0.1892132132 |
- |
- |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 6 |
Hb_001863_480 |
0.1927923418 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 7 |
Hb_000539_030 |
0.193317488 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 8 |
Hb_000212_350 |
0.1992180705 |
- |
- |
glucosamine-fructose-6-phosphate aminotransferase, putative [Ricinus communis] |
| 9 |
Hb_164926_030 |
0.2003454683 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 10 |
Hb_007831_020 |
0.2017225968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002471_100 |
0.2037567146 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_080477_050 |
0.2041854131 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 13 |
Hb_005375_020 |
0.2079673339 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 1-like [Nicotiana sylvestris] |
| 14 |
Hb_003849_100 |
0.2086530914 |
- |
- |
PREDICTED: putative transcription elongation factor SPT5 homolog 1 [Populus euphratica] |
| 15 |
Hb_028487_030 |
0.2088685634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001587_070 |
0.2090612282 |
desease resistance |
Gene Name: NB-ARC |
Nbs-lrr resistance protein, putative [Theobroma cacao] |
| 17 |
Hb_001439_170 |
0.2119409793 |
- |
- |
PREDICTED: pectinesterase 31-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001828_080 |
0.2141404085 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase family protein [Populus trichocarpa] |
| 19 |
Hb_000169_140 |
0.2146578683 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 20 |
Hb_000258_330 |
0.2149821948 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |