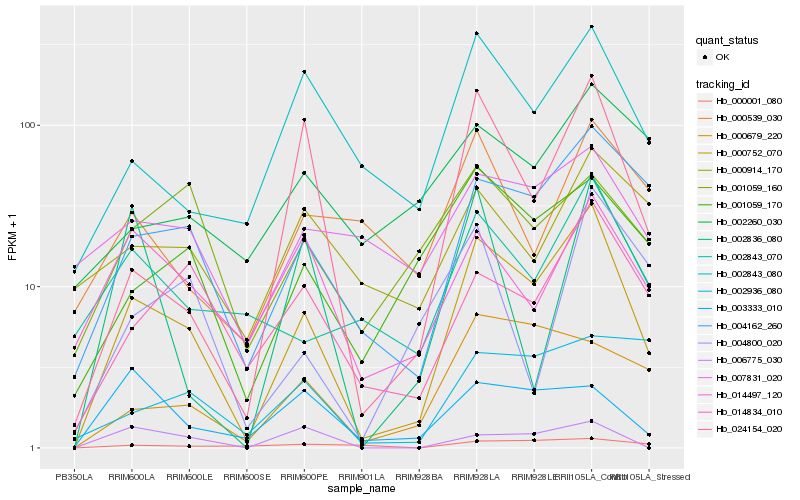
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000752_070 |
0.0 |
- |
- |
PREDICTED: mavicyanin-like [Jatropha curcas] |
| 2 |
Hb_004162_260 |
0.2092368402 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_024154_020 |
0.2443412442 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 4 |
Hb_014834_010 |
0.2449785248 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4 [Jatropha curcas] |
| 5 |
Hb_002843_080 |
0.2463692994 |
- |
- |
SAUR family protein [Theobroma cacao] |
| 6 |
Hb_001059_170 |
0.25096782 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 7 |
Hb_002843_070 |
0.2589348151 |
- |
- |
unknown [Lotus japonicus] |
| 8 |
Hb_014497_120 |
0.2686027223 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 9 |
Hb_000679_220 |
0.2717576594 |
- |
- |
PREDICTED: uncharacterized protein LOC105637638 [Jatropha curcas] |
| 10 |
Hb_007831_020 |
0.284901273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000539_030 |
0.2872408742 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 12 |
Hb_002836_080 |
0.2891833462 |
- |
- |
- |
| 13 |
Hb_000001_080 |
0.2913015113 |
- |
- |
- |
| 14 |
Hb_001059_160 |
0.2915744378 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 15 |
Hb_004800_020 |
0.2917367623 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL4-like [Jatropha curcas] |
| 16 |
Hb_002260_030 |
0.2926161636 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 17 |
Hb_006775_030 |
0.2926671058 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 18 |
Hb_003333_010 |
0.2938765268 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 19 |
Hb_000914_170 |
0.2947148302 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 20 |
Hb_002936_080 |
0.3002023179 |
- |
- |
Protein bem46, putative [Ricinus communis] |