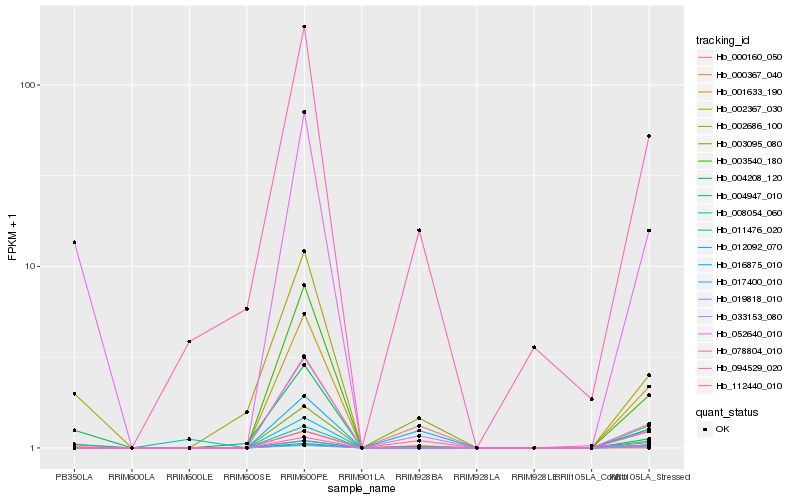
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001633_190 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 2 |
Hb_052640_010 |
0.2069548716 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004947_010 |
0.2386648036 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 4 |
Hb_033153_080 |
0.2514964969 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 5 |
Hb_016875_010 |
0.2548327594 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_002686_100 |
0.2741414062 |
- |
- |
- |
| 7 |
Hb_094529_020 |
0.2753399006 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 8 |
Hb_000160_050 |
0.275391927 |
- |
- |
hypothetical protein JCGZ_02516 [Jatropha curcas] |
| 9 |
Hb_008054_060 |
0.2860499475 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 10 |
Hb_078804_010 |
0.2895453628 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_004208_120 |
0.2961681022 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 12 |
Hb_002367_030 |
0.311829982 |
- |
- |
hypothetical protein POPTR_0001s30725g [Populus trichocarpa] |
| 13 |
Hb_017400_010 |
0.3154401125 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_019818_010 |
0.3223632925 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 15 |
Hb_000367_040 |
0.3265657916 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_112440_010 |
0.331098708 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 17 |
Hb_003540_180 |
0.3319329894 |
- |
- |
- |
| 18 |
Hb_012092_070 |
0.3327237978 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 19 |
Hb_011476_020 |
0.3339752179 |
- |
- |
PREDICTED: glutamate dehydrogenase 2-like [Jatropha curcas] |
| 20 |
Hb_003095_080 |
0.3392434846 |
- |
- |
- |