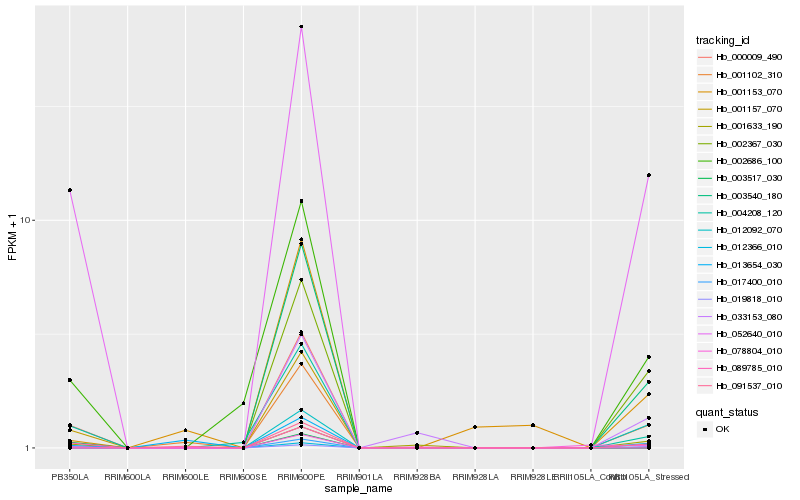
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052640_010 |
0.0 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002686_100 |
0.1748714684 |
- |
- |
- |
| 3 |
Hb_004208_120 |
0.1811724838 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 4 |
Hb_001633_190 |
0.2069548716 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 5 |
Hb_002367_030 |
0.2627774421 |
- |
- |
hypothetical protein POPTR_0001s30725g [Populus trichocarpa] |
| 6 |
Hb_003540_180 |
0.2675512468 |
- |
- |
- |
| 7 |
Hb_017400_010 |
0.2822957906 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_089785_010 |
0.2848225727 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 9 |
Hb_012366_010 |
0.2856895542 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 10 |
Hb_001157_070 |
0.288648059 |
- |
- |
- |
| 11 |
Hb_000009_490 |
0.2896361923 |
- |
- |
hypothetical protein POPTR_0007s12040g [Populus trichocarpa] |
| 12 |
Hb_001102_310 |
0.2901175852 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 13 |
Hb_019818_010 |
0.2964232291 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 14 |
Hb_091537_010 |
0.3059318948 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001153_070 |
0.3077928572 |
- |
- |
- |
| 16 |
Hb_003517_030 |
0.3088180745 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 5 isoform X2 [Eucalyptus grandis] |
| 17 |
Hb_013654_030 |
0.312748523 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 18 |
Hb_033153_080 |
0.3129250813 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 19 |
Hb_012092_070 |
0.3139680222 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 20 |
Hb_078804_010 |
0.3179288855 |
- |
- |
kinase, putative [Ricinus communis] |