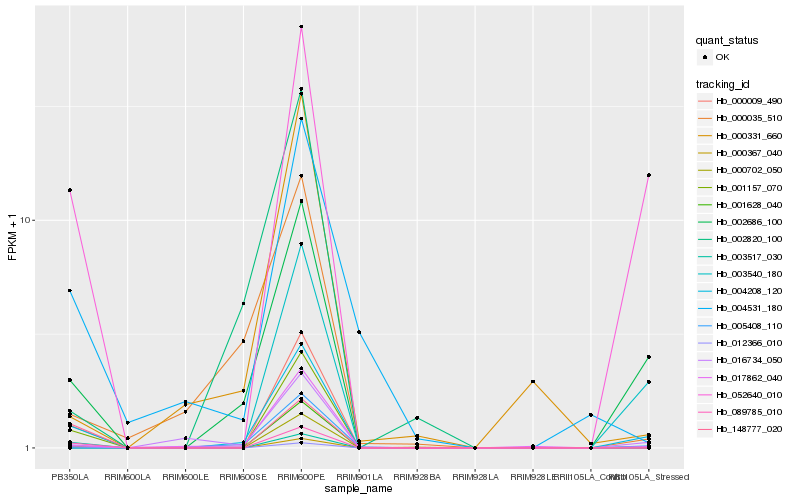
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004208_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 2 |
Hb_002686_100 |
0.1053177052 |
- |
- |
- |
| 3 |
Hb_052640_010 |
0.1811724838 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001157_070 |
0.2016944989 |
- |
- |
- |
| 5 |
Hb_000009_490 |
0.2021239133 |
- |
- |
hypothetical protein POPTR_0007s12040g [Populus trichocarpa] |
| 6 |
Hb_089785_010 |
0.2127671864 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 7 |
Hb_012366_010 |
0.2154982919 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 8 |
Hb_000702_050 |
0.2353474675 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_017862_040 |
0.2407036416 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 3-like [Prunus mume] |
| 10 |
Hb_016734_050 |
0.2454070191 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 11 |
Hb_003517_030 |
0.2606144203 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 5 isoform X2 [Eucalyptus grandis] |
| 12 |
Hb_001628_040 |
0.2664348856 |
- |
- |
PREDICTED: beta-galactosidase 13-like [Populus euphratica] |
| 13 |
Hb_000367_040 |
0.2704321404 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_005408_110 |
0.2717312881 |
- |
- |
purine permease, putative [Ricinus communis] |
| 15 |
Hb_148777_020 |
0.2754575098 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 isoform X1 [Populus euphratica] |
| 16 |
Hb_000035_510 |
0.2774649247 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 17 |
Hb_002820_100 |
0.2780048598 |
- |
- |
hypothetical protein OsI_33031 [Oryza sativa Indica Group] |
| 18 |
Hb_004531_180 |
0.2781168332 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_003540_180 |
0.2789113862 |
- |
- |
- |
| 20 |
Hb_000331_660 |
0.2816293731 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |