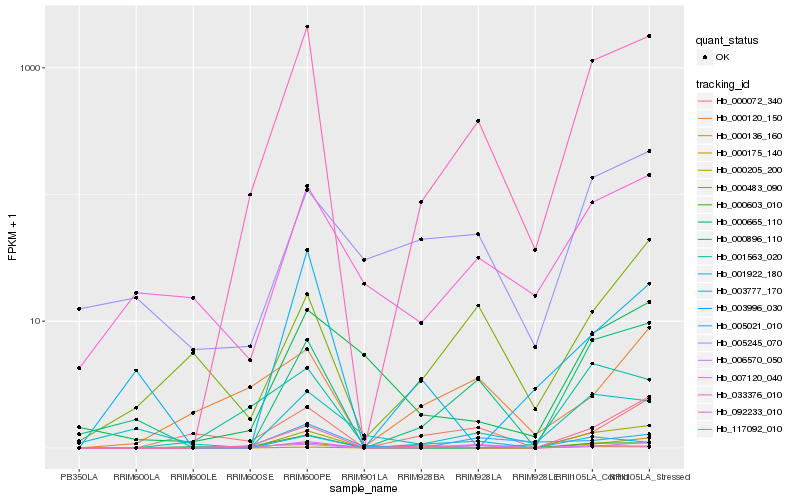
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092233_010 |
0.0 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 2 |
Hb_000175_140 |
0.207773793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000896_110 |
0.2126280156 |
- |
- |
- |
| 4 |
Hb_000205_200 |
0.2458174905 |
- |
- |
- |
| 5 |
Hb_033376_010 |
0.272159217 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 6 |
Hb_005021_010 |
0.2785096728 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 7 |
Hb_000072_340 |
0.28738662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000665_110 |
0.2909216804 |
- |
- |
Ribosomal protein S11-beta [Theobroma cacao] |
| 9 |
Hb_000136_160 |
0.2959753878 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_007120_040 |
0.2963087745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006570_050 |
0.3018460418 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 12 |
Hb_003777_170 |
0.3022445116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005245_070 |
0.3026229656 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 14 |
Hb_000603_010 |
0.3029577138 |
- |
- |
PREDICTED: transmembrane protein 87A-like [Nelumbo nucifera] |
| 15 |
Hb_117092_010 |
0.3031633596 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 16 |
Hb_000483_090 |
0.3035584342 |
- |
- |
- |
| 17 |
Hb_001922_180 |
0.3057819588 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 18 |
Hb_001563_020 |
0.3060390114 |
transcription factor |
TF Family: TRAF |
aberrant large forked product, putative [Ricinus communis] |
| 19 |
Hb_000120_150 |
0.3068612072 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 20 |
Hb_003996_030 |
0.3089327996 |
- |
- |
- |