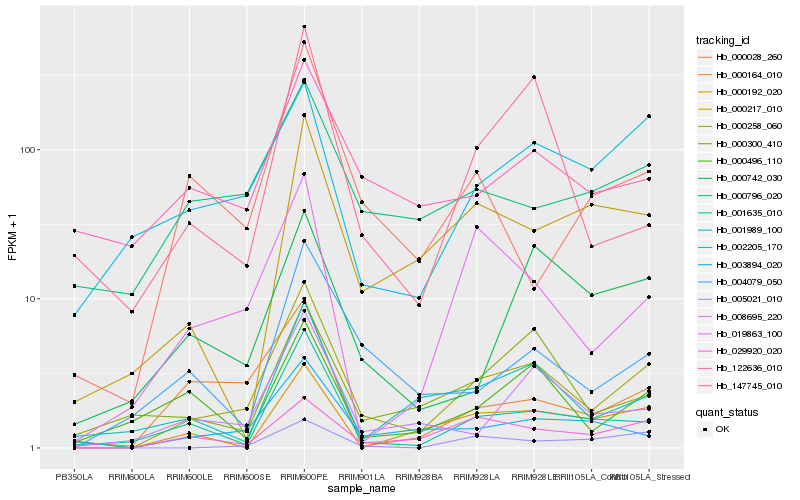
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001635_010 |
0.1982834509 |
- |
- |
- |
| 3 |
Hb_008695_220 |
0.2041941031 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 4 |
Hb_003894_020 |
0.2117525763 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 5 |
Hb_001989_100 |
0.2159927653 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 6 |
Hb_000796_020 |
0.2221681252 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 7 |
Hb_019863_100 |
0.2226835414 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_122636_010 |
0.2261782084 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 9 |
Hb_004079_050 |
0.226194938 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 10 |
Hb_147745_010 |
0.2294449995 |
- |
- |
PREDICTED: uncharacterized protein LOC105639149 [Jatropha curcas] |
| 11 |
Hb_029920_020 |
0.2298481162 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 12 |
Hb_000164_010 |
0.230004478 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 13 |
Hb_002205_170 |
0.2323763028 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 14 |
Hb_000217_010 |
0.2337442857 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 15 |
Hb_000028_260 |
0.2371703428 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 16 |
Hb_000496_110 |
0.2372362127 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 17 |
Hb_000300_410 |
0.2394832957 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 18 |
Hb_000742_030 |
0.2446505006 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 19 |
Hb_005021_010 |
0.2467072739 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 20 |
Hb_000192_020 |
0.2497072876 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |