| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
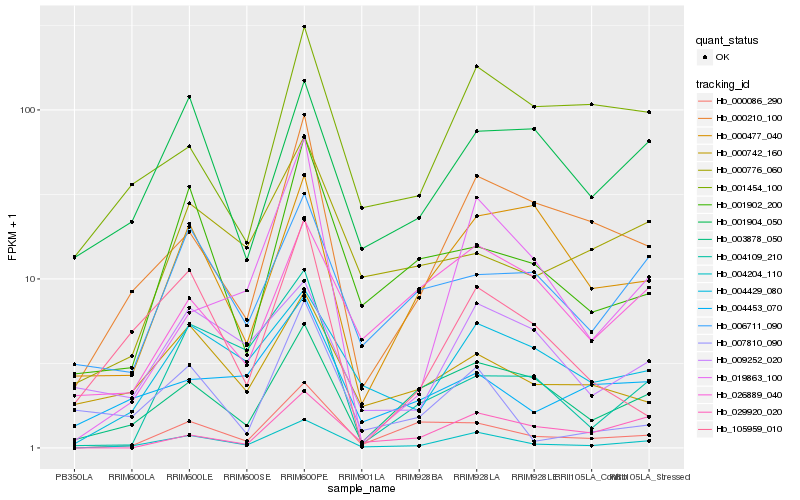
Hb_004204_110 |
0.0 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 2 |
Hb_004429_080 |
0.1951831334 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_003878_050 |
0.198316797 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 4 |
Hb_000086_290 |
0.2012389556 |
- |
- |
PREDICTED: TBC1 domain family member 15-like [Jatropha curcas] |
| 5 |
Hb_000210_100 |
0.2045569662 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 6 |
Hb_001902_200 |
0.2197385033 |
- |
- |
PREDICTED: tubulin beta-1 chain [Eucalyptus grandis] |
| 7 |
Hb_006711_090 |
0.2274992196 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 8 |
Hb_019863_100 |
0.2277038693 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_009252_020 |
0.2304668115 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 10 |
Hb_001904_050 |
0.2312388988 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 11 |
Hb_000742_160 |
0.2323865705 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 12 |
Hb_105959_010 |
0.2342572826 |
- |
- |
asparagine synthetase, putative [Ricinus communis] |
| 13 |
Hb_000776_060 |
0.2393006396 |
- |
- |
histone H4 [Zea mays] |
| 14 |
Hb_004453_070 |
0.2406284713 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_029920_020 |
0.2419283213 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 16 |
Hb_000477_040 |
0.2426396571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001454_100 |
0.2427747461 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 18 |
Hb_007810_090 |
0.2434618001 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 19 |
Hb_004109_210 |
0.2447180079 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_026889_040 |
0.2457378685 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |