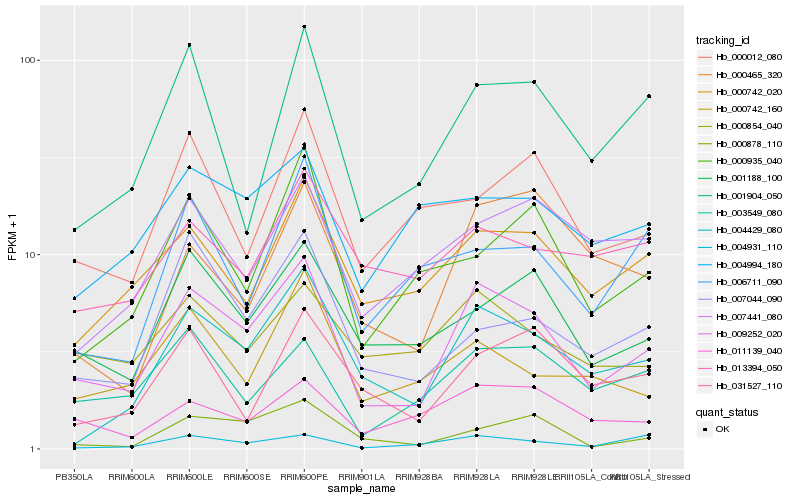
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009252_020 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 2 |
Hb_004429_080 |
0.148472855 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001188_100 |
0.1640844784 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 4 |
Hb_000742_020 |
0.1712096504 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 5 |
Hb_003549_080 |
0.1755599411 |
- |
- |
PREDICTED: importin subunit alpha-4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001904_050 |
0.1773472122 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 7 |
Hb_011139_040 |
0.1782179268 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000854_040 |
0.1788943547 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 9 |
Hb_007044_090 |
0.1811299964 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 10 |
Hb_000878_110 |
0.1837117797 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000742_160 |
0.1864536283 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 12 |
Hb_000935_040 |
0.1888725706 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_000012_080 |
0.1918062775 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 14 |
Hb_004931_110 |
0.1929445352 |
- |
- |
inorganic phosphate transporter, putative [Ricinus communis] |
| 15 |
Hb_004994_180 |
0.1952000176 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 16 |
Hb_007441_080 |
0.1956030217 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 17 |
Hb_000465_320 |
0.1994913919 |
- |
- |
PREDICTED: nudix hydrolase 8 [Jatropha curcas] |
| 18 |
Hb_013394_050 |
0.2001287175 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 19 |
Hb_031527_110 |
0.2002385836 |
- |
- |
PREDICTED: probable glutathione S-transferase parC [Jatropha curcas] |
| 20 |
Hb_006711_090 |
0.2005172629 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |