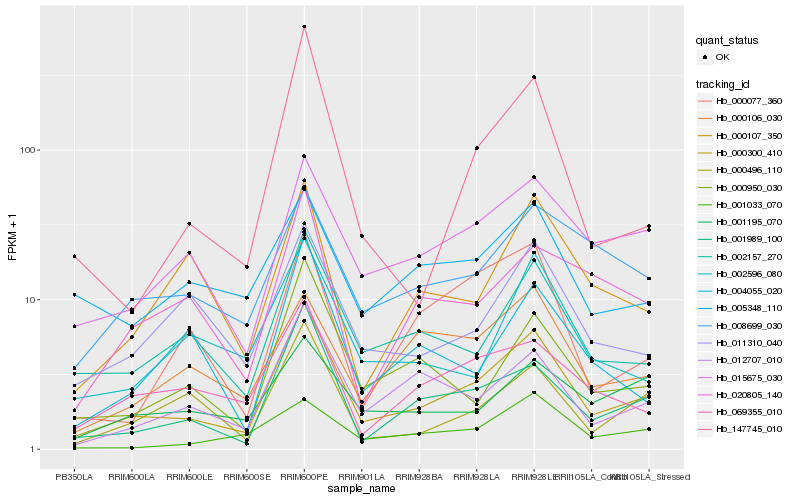
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_410 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s05860g [Populus trichocarpa] |
| 2 |
Hb_001989_100 |
0.1457248099 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 3 |
Hb_002157_270 |
0.1490611231 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 4 |
Hb_012707_010 |
0.1603002729 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 5 |
Hb_015675_030 |
0.1606995312 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000950_030 |
0.1702336576 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 7 |
Hb_011310_040 |
0.1729193728 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 8 |
Hb_000496_110 |
0.1732411256 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 9 |
Hb_069355_010 |
0.1757165295 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |
| 10 |
Hb_147745_010 |
0.1813207264 |
- |
- |
PREDICTED: uncharacterized protein LOC105639149 [Jatropha curcas] |
| 11 |
Hb_002596_080 |
0.1854297574 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 12 |
Hb_000107_350 |
0.1870096439 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 13 |
Hb_008699_030 |
0.1873768715 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 14 |
Hb_020805_140 |
0.1893589401 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 15 |
Hb_000077_360 |
0.193112582 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_001195_070 |
0.1943415483 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 17 |
Hb_004055_020 |
0.1944169199 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 18 |
Hb_005348_110 |
0.1960727294 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 19 |
Hb_000106_030 |
0.1960853682 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_001033_070 |
0.1966361127 |
- |
- |
seryl-tRNA synthetase, putative [Ricinus communis] |