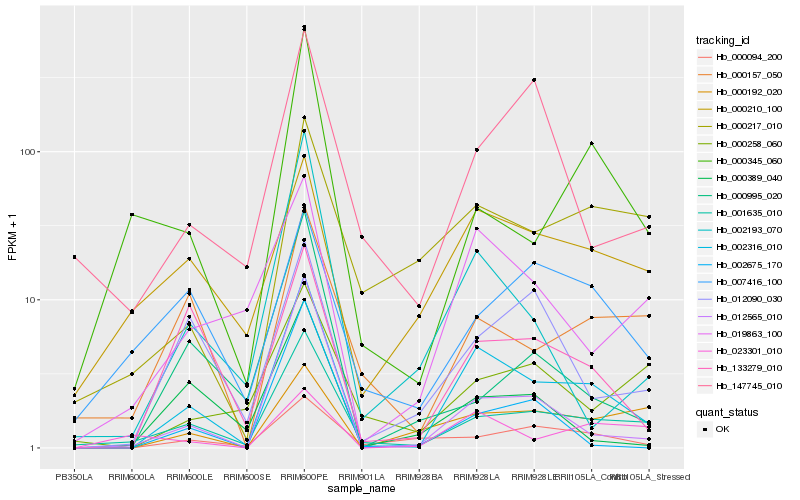
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002316_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 2 |
Hb_001635_010 |
0.189421735 |
- |
- |
- |
| 3 |
Hb_012565_010 |
0.2211798685 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 4 |
Hb_133279_010 |
0.2350428971 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 5 |
Hb_019863_100 |
0.2362864851 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_002193_070 |
0.2445861467 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000157_050 |
0.2605114374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000345_060 |
0.2634295239 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 9 |
Hb_002675_170 |
0.264767273 |
- |
- |
hypothetical protein JCGZ_06235 [Jatropha curcas] |
| 10 |
Hb_000210_100 |
0.2649852345 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 11 |
Hb_000389_040 |
0.2663092077 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g03250-like [Jatropha curcas] |
| 12 |
Hb_000094_200 |
0.2668118953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000217_010 |
0.2741357127 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 14 |
Hb_012090_030 |
0.2741994898 |
- |
- |
PREDICTED: autophagy-related protein 8C isoform X2 [Phoenix dactylifera] |
| 15 |
Hb_000192_020 |
0.2855481303 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 16 |
Hb_000258_060 |
0.2895750367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023301_010 |
0.2898725404 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 18 |
Hb_147745_010 |
0.2905348172 |
- |
- |
PREDICTED: uncharacterized protein LOC105639149 [Jatropha curcas] |
| 19 |
Hb_000995_020 |
0.2906368457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007416_100 |
0.29696572 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |