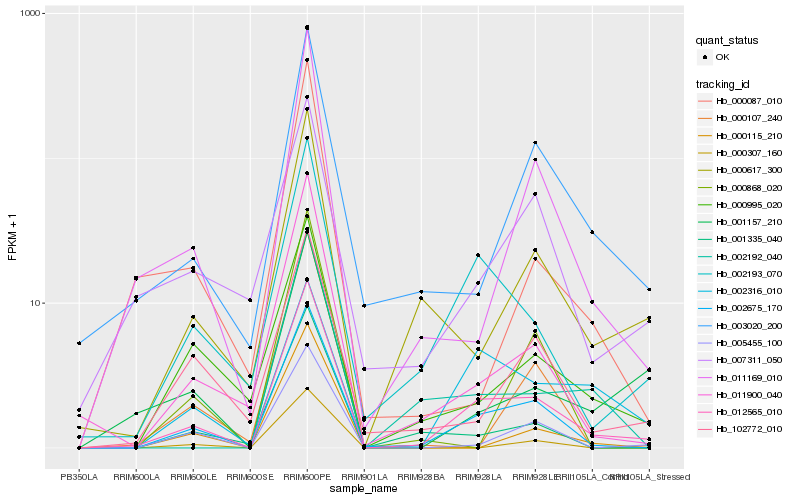
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012565_010 |
0.0 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 2 |
Hb_002675_170 |
0.1016872835 |
- |
- |
hypothetical protein JCGZ_06235 [Jatropha curcas] |
| 3 |
Hb_002193_070 |
0.1511842248 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_011900_040 |
0.1746752357 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 5 |
Hb_011169_010 |
0.181179138 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 6 |
Hb_005455_100 |
0.1852821443 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_000617_300 |
0.1884554718 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 8 |
Hb_001335_040 |
0.1906561858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003020_200 |
0.1908737345 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 10 |
Hb_000995_020 |
0.1924685439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001157_210 |
0.2006753424 |
- |
- |
hypothetical protein CISIN_1g038260mg [Citrus sinensis] |
| 12 |
Hb_102772_010 |
0.2046191379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002192_040 |
0.2053337971 |
- |
- |
- |
| 14 |
Hb_007311_050 |
0.2135913804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000115_210 |
0.2143906702 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 16 |
Hb_000107_240 |
0.2176381554 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 17 |
Hb_002316_010 |
0.2211798685 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 18 |
Hb_000868_020 |
0.2240805919 |
- |
- |
- |
| 19 |
Hb_000087_010 |
0.2244690149 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 20 |
Hb_000307_160 |
0.225078432 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |