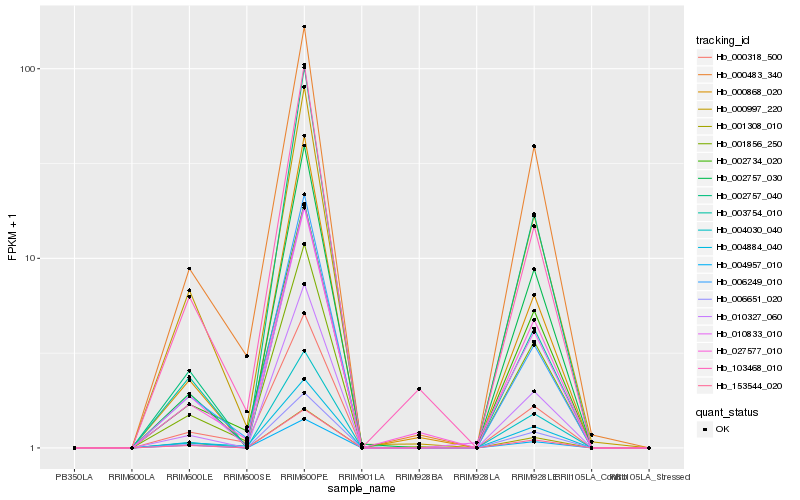
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010833_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |
| 2 |
Hb_153544_020 |
0.0451444395 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP12 [Jatropha curcas] |
| 3 |
Hb_003754_010 |
0.0594755085 |
- |
- |
hypothetical protein POPTR_0019s09950g [Populus trichocarpa] |
| 4 |
Hb_006651_020 |
0.0596619625 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 5 |
Hb_004957_010 |
0.0632725917 |
- |
- |
PREDICTED: probable aquaporin NIP-type [Prunus mume] |
| 6 |
Hb_000997_220 |
0.0639923614 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA14-like [Jatropha curcas] |
| 7 |
Hb_010327_060 |
0.0667949899 |
- |
- |
hypothetical protein B456_001G246300 [Gossypium raimondii] |
| 8 |
Hb_004030_040 |
0.0716559177 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.8 [Jatropha curcas] |
| 9 |
Hb_002757_030 |
0.072629349 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_006249_010 |
0.075305729 |
- |
- |
hypothetical protein JCGZ_13152 [Jatropha curcas] |
| 11 |
Hb_001856_250 |
0.0776929456 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 12 |
Hb_000318_500 |
0.0804209373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000483_340 |
0.083842948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000868_020 |
0.0839968755 |
- |
- |
- |
| 15 |
Hb_002734_020 |
0.084547204 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 16 |
Hb_001308_010 |
0.0858214294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004884_040 |
0.0878291498 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 18 |
Hb_002757_040 |
0.0882398532 |
- |
- |
- |
| 19 |
Hb_027577_010 |
0.090658863 |
- |
- |
PREDICTED: uncharacterized protein LOC104779928 [Camelina sativa] |
| 20 |
Hb_103468_010 |
0.0914957059 |
- |
- |
protein binding protein, putative [Ricinus communis] |