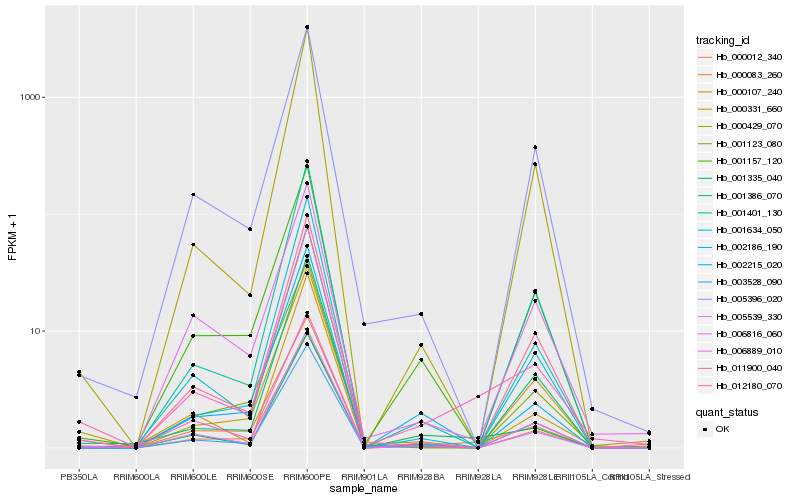
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011900_040 |
0.0 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 2 |
Hb_000083_260 |
0.1059170529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001335_040 |
0.1091993234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005539_330 |
0.1184714827 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 5 |
Hb_000331_660 |
0.1279629356 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |
| 6 |
Hb_000012_340 |
0.1323045535 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 7 |
Hb_000429_070 |
0.1340752506 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 8 |
Hb_012180_070 |
0.1345430138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005396_020 |
0.1354410503 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 10 |
Hb_001401_130 |
0.1367590541 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 11 |
Hb_006889_010 |
0.1384232577 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 12 |
Hb_001386_070 |
0.1384643857 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_003528_090 |
0.1403082504 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 14 |
Hb_001634_050 |
0.1403179996 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 12 [Jatropha curcas] |
| 15 |
Hb_001157_120 |
0.1412609367 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 16 |
Hb_001123_080 |
0.1454252372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002186_190 |
0.1476639304 |
- |
- |
- |
| 18 |
Hb_002215_020 |
0.1478863095 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 19 |
Hb_006816_060 |
0.1479843429 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 20 |
Hb_000107_240 |
0.1497398495 |
- |
- |
organic anion transporter, putative [Ricinus communis] |