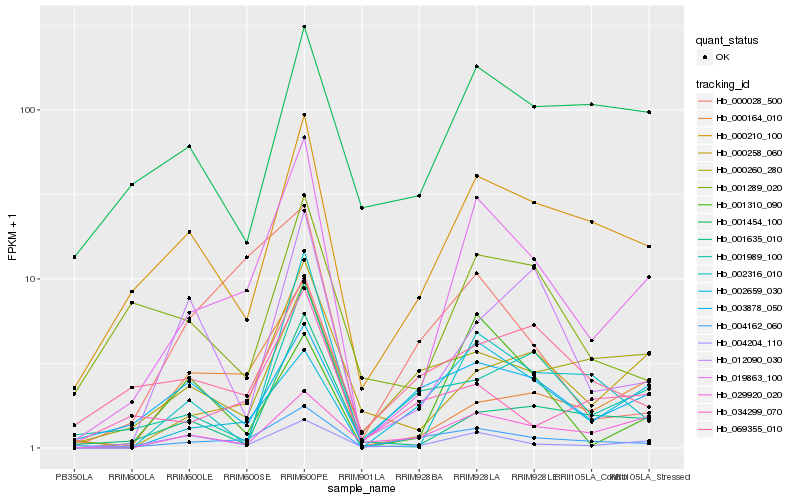
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019863_100 |
0.0 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_000210_100 |
0.215071916 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 3 |
Hb_004162_060 |
0.2224195779 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000258_060 |
0.2226835414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004204_110 |
0.2277038693 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 6 |
Hb_002316_010 |
0.2362864851 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 7 |
Hb_000164_010 |
0.2494457676 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 8 |
Hb_001635_010 |
0.2501650694 |
- |
- |
- |
| 9 |
Hb_029920_020 |
0.2559120983 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 10 |
Hb_034299_070 |
0.2624631128 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 11 |
Hb_002659_030 |
0.2663412719 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 12 |
Hb_001289_020 |
0.2685749 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 13 |
Hb_001989_100 |
0.2688830606 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 14 |
Hb_003878_050 |
0.2717853086 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 15 |
Hb_000260_280 |
0.2768323064 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 16 |
Hb_001310_090 |
0.2788769411 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 17 |
Hb_001454_100 |
0.2797791718 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 18 |
Hb_012090_030 |
0.2804416535 |
- |
- |
PREDICTED: autophagy-related protein 8C isoform X2 [Phoenix dactylifera] |
| 19 |
Hb_000028_500 |
0.2842408756 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_069355_010 |
0.2844771271 |
- |
- |
NBS-LRR resistance protein RGH2 [Manihot esculenta] |