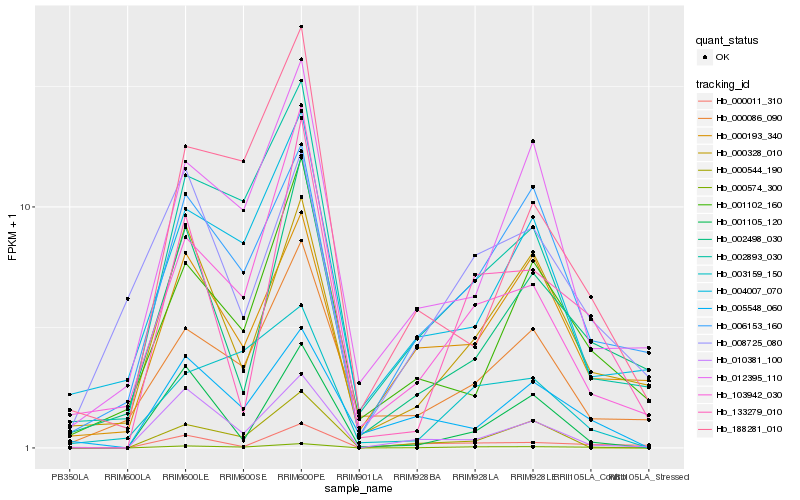
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_300 |
0.0 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 2 |
Hb_133279_010 |
0.1981609438 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 3 |
Hb_002893_030 |
0.2003276546 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 4 |
Hb_006153_160 |
0.2070985579 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X4 [Jatropha curcas] |
| 5 |
Hb_005548_060 |
0.2080462676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003159_150 |
0.2108967486 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 8 [Jatropha curcas] |
| 7 |
Hb_103942_030 |
0.2169402322 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_010381_100 |
0.2205117237 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 9 |
Hb_000011_310 |
0.2218254478 |
- |
- |
PREDICTED: uncharacterized protein LOC105631327 [Jatropha curcas] |
| 10 |
Hb_000328_010 |
0.225111947 |
- |
- |
PREDICTED: QWRF motif-containing protein 8-like [Jatropha curcas] |
| 11 |
Hb_000086_090 |
0.2298334464 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 12 |
Hb_012395_110 |
0.2309981743 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 13 |
Hb_188281_010 |
0.2310877613 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 14 |
Hb_000193_340 |
0.2314939821 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001102_160 |
0.2316627416 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 16 |
Hb_002498_030 |
0.2330498955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000544_190 |
0.2343061181 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 18 |
Hb_004007_070 |
0.2377731948 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 19 |
Hb_001105_120 |
0.238903188 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 20 |
Hb_008725_080 |
0.2397824046 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |