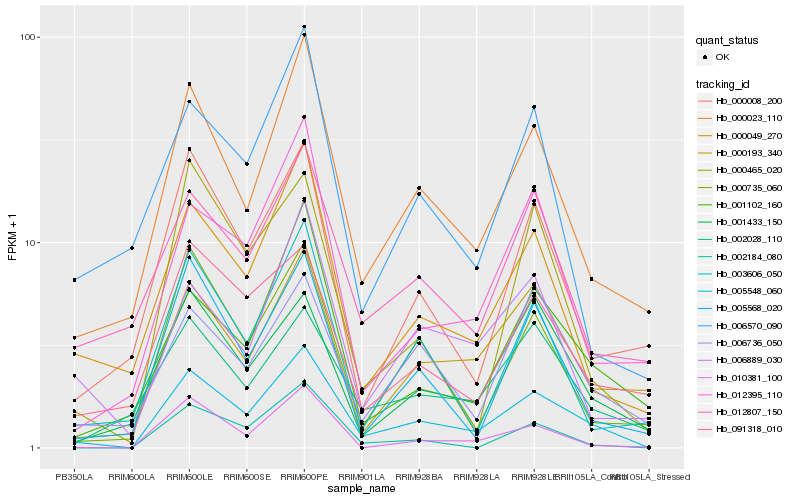
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005548_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000023_110 |
0.1438670892 |
- |
- |
PREDICTED: probable methyltransferase PMT21 [Jatropha curcas] |
| 3 |
Hb_000049_270 |
0.1674933164 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase ALE2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005568_020 |
0.1840788569 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000735_060 |
0.1850036714 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 6 |
Hb_000193_340 |
0.1852543927 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001433_150 |
0.1887874231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006736_050 |
0.1892099411 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 9 |
Hb_010381_100 |
0.1896072846 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 10 |
Hb_001102_160 |
0.1914213134 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 11 |
Hb_012807_150 |
0.1922555066 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 12 |
Hb_002028_110 |
0.192876509 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 13 |
Hb_002184_080 |
0.1941058978 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 14 |
Hb_003606_050 |
0.1945546678 |
- |
- |
PREDICTED: beta-galactosidase 10 [Jatropha curcas] |
| 15 |
Hb_006889_030 |
0.1948123203 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000465_020 |
0.1956304077 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 17 |
Hb_091318_010 |
0.1957435065 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 18 |
Hb_012395_110 |
0.196291169 |
- |
- |
PREDICTED: peroxygenase [Jatropha curcas] |
| 19 |
Hb_000008_200 |
0.1973808793 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 20 |
Hb_006570_090 |
0.197595692 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |