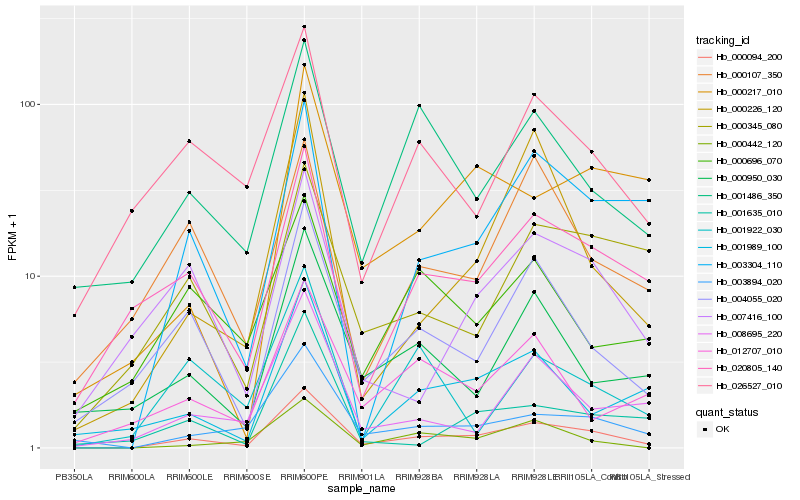
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003894_020 |
0.1792652524 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 3 |
Hb_004055_020 |
0.1850379908 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 4 |
Hb_020805_140 |
0.193667119 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 5 |
Hb_003304_110 |
0.1971957821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000442_120 |
0.1997523896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008695_220 |
0.2053478029 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 8 |
Hb_001486_350 |
0.2088460177 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001989_100 |
0.2101181989 |
- |
- |
PREDICTED: uncharacterized protein LOC105636214 [Jatropha curcas] |
| 10 |
Hb_026527_010 |
0.2119433008 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 11 |
Hb_000950_030 |
0.2125586629 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 12 |
Hb_000226_120 |
0.2176998972 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012707_010 |
0.2179287647 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 14 |
Hb_007416_100 |
0.2219295882 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 5 [Jatropha curcas] |
| 15 |
Hb_000345_080 |
0.230115723 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 16 |
Hb_000217_010 |
0.2326873166 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 17 |
Hb_000107_350 |
0.2338564224 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 18 |
Hb_000696_070 |
0.2342748097 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 19 |
Hb_001635_010 |
0.2352225631 |
- |
- |
- |
| 20 |
Hb_001922_030 |
0.2358619892 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |