| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
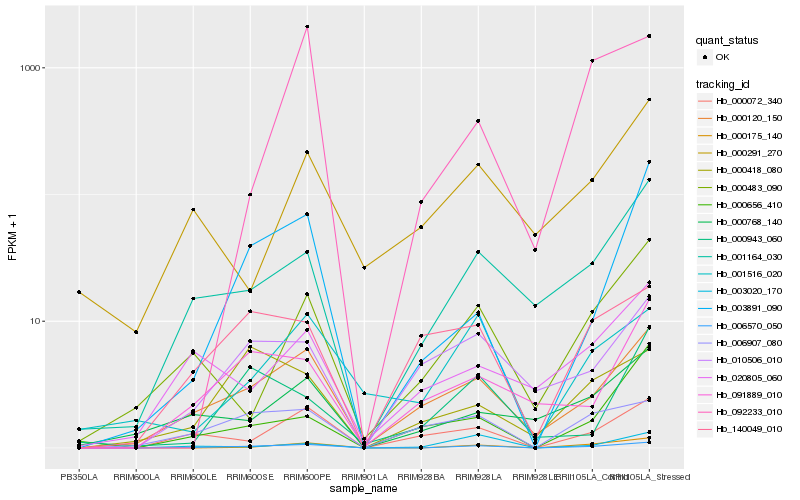
Hb_006570_050 |
0.0 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 2 |
Hb_000120_150 |
0.1961499775 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 3 |
Hb_000175_140 |
0.2047959205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001164_030 |
0.2360664727 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 5 |
Hb_000072_340 |
0.2369540991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003891_090 |
0.2596971635 |
- |
- |
- |
| 7 |
Hb_006907_080 |
0.2620158993 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 8 |
Hb_000483_090 |
0.2692685773 |
- |
- |
- |
| 9 |
Hb_091889_010 |
0.2725991011 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001516_020 |
0.2733881576 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-9-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000768_140 |
0.2752924885 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_010506_010 |
0.2763081569 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_140049_010 |
0.2805159866 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 14 |
Hb_003020_170 |
0.2831664982 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 15 |
Hb_000418_080 |
0.2898141878 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 16 |
Hb_020805_060 |
0.2907156896 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 17 |
Hb_092233_010 |
0.3018460418 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 18 |
Hb_000656_410 |
0.3032172365 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 19 |
Hb_000943_060 |
0.3084746548 |
- |
- |
PREDICTED: uncharacterized protein LOC105635589 [Jatropha curcas] |
| 20 |
Hb_000291_270 |
0.3096972171 |
- |
- |
hypothetical protein JCGZ_12006 [Jatropha curcas] |