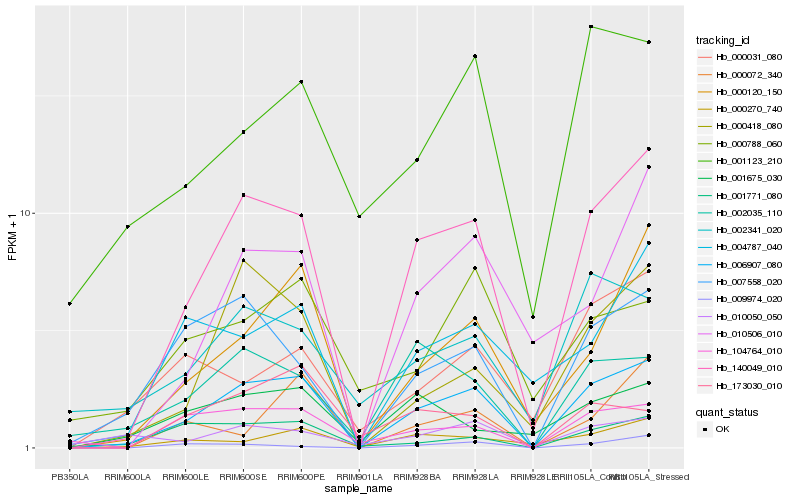
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006907_080 |
0.0 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 2 |
Hb_140049_010 |
0.0851991089 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 3 |
Hb_000120_150 |
0.1975733222 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 4 |
Hb_007558_020 |
0.2042281158 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 5 |
Hb_000418_080 |
0.21022697 |
- |
- |
PREDICTED: uncharacterized protein LOC105124652 [Populus euphratica] |
| 6 |
Hb_104764_010 |
0.2147474247 |
- |
- |
hypothetical protein JCGZ_16024 [Jatropha curcas] |
| 7 |
Hb_002035_110 |
0.219144102 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 8 |
Hb_010506_010 |
0.2217252814 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_000270_740 |
0.2251876646 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 10 |
Hb_004787_040 |
0.2259892445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001123_210 |
0.2304444166 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 12 |
Hb_002341_020 |
0.2308610421 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 13 |
Hb_001771_080 |
0.2344331493 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Amborella trichopoda] |
| 14 |
Hb_173030_010 |
0.2398545884 |
- |
- |
hypothetical protein RCOM_1321060 [Ricinus communis] |
| 15 |
Hb_010050_050 |
0.2414372843 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_000031_080 |
0.242260347 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 17 |
Hb_001675_030 |
0.2454932484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000072_340 |
0.2522115141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009974_020 |
0.2529822461 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 20 |
Hb_000788_060 |
0.2531469099 |
- |
- |
PREDICTED: uncharacterized protein LOC105629705 [Jatropha curcas] |