| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
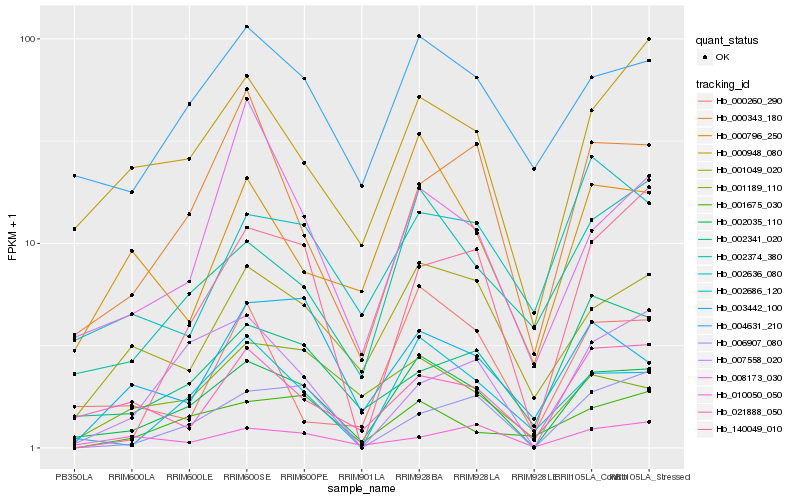
Hb_002035_110 |
0.0 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 2 |
Hb_002686_120 |
0.1661019533 |
- |
- |
short-chain type dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_001049_020 |
0.1765249423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004631_210 |
0.1767161369 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 5 |
Hb_000948_080 |
0.1860341929 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 6 |
Hb_002374_380 |
0.1897438323 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_000343_180 |
0.1974615937 |
- |
- |
NDWp3 [Podospora anserina] |
| 8 |
Hb_021888_050 |
0.2032045021 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 9 |
Hb_000796_250 |
0.2057703171 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 10 |
Hb_002341_020 |
0.2100718024 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 11 |
Hb_001675_030 |
0.2115202467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008173_030 |
0.2155039363 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 13 |
Hb_140049_010 |
0.2172673329 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 14 |
Hb_001189_110 |
0.218386339 |
- |
- |
NAC transcription factor 066 [Jatropha curcas] |
| 15 |
Hb_006907_080 |
0.219144102 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 16 |
Hb_000260_290 |
0.2208501266 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003442_100 |
0.2251937355 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 18 |
Hb_007558_020 |
0.225967051 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 19 |
Hb_002636_080 |
0.2261899667 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 3-like [Jatropha curcas] |
| 20 |
Hb_010050_050 |
0.2283704982 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |