| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
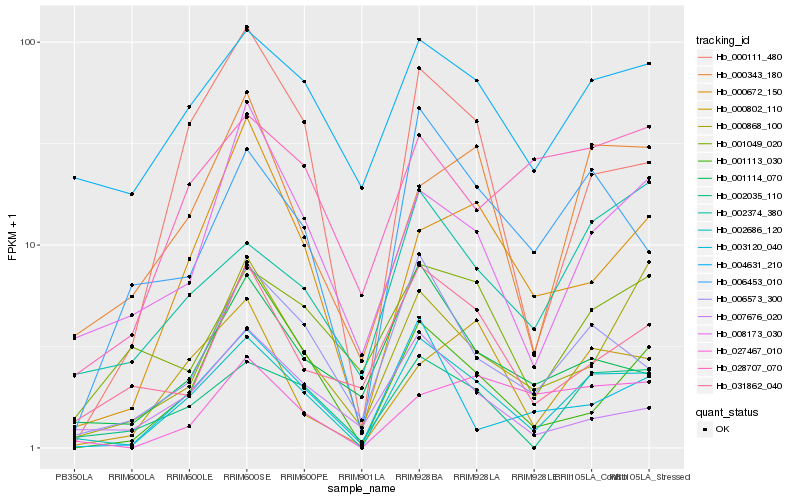
Hb_002686_120 |
0.0 |
- |
- |
short-chain type dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_002035_110 |
0.1661019533 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 3 |
Hb_000111_480 |
0.1813836529 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 4 |
Hb_006573_300 |
0.1828257079 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_004631_210 |
0.1923991831 |
- |
- |
hypothetical protein POPTR_0007s05070g [Populus trichocarpa] |
| 6 |
Hb_006453_010 |
0.205414826 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 7 |
Hb_000343_180 |
0.2082667778 |
- |
- |
NDWp3 [Podospora anserina] |
| 8 |
Hb_001114_070 |
0.2088184923 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 9 |
Hb_003120_040 |
0.2116856498 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 10 |
Hb_008173_030 |
0.2158722924 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 11 |
Hb_002374_380 |
0.2170778616 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_000868_100 |
0.21781896 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 13 |
Hb_000802_110 |
0.2228821961 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 14 |
Hb_031862_040 |
0.2252227253 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 15 |
Hb_007676_020 |
0.2259569097 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000672_150 |
0.2275750818 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 17 |
Hb_028707_070 |
0.2318149116 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_027467_010 |
0.2325125033 |
- |
- |
hypothetical protein PRUPE_ppa004106mg [Prunus persica] |
| 19 |
Hb_001049_020 |
0.2325280822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001113_030 |
0.2327535068 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |