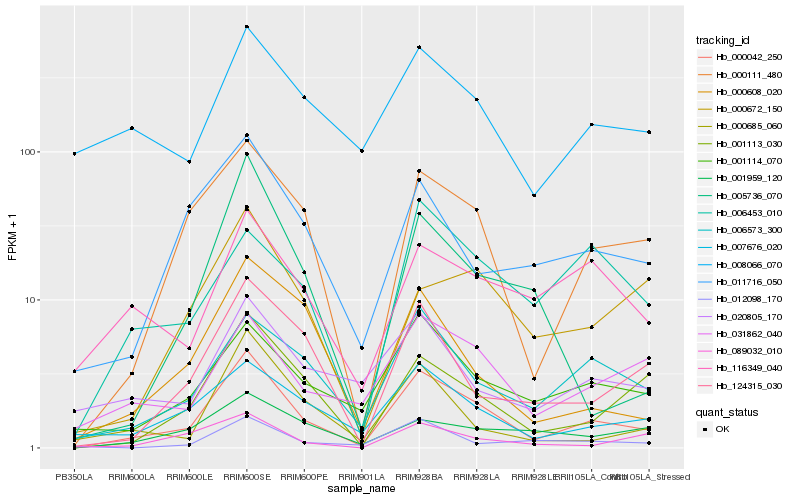
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000111_480 |
0.1739021541 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 3 |
Hb_006573_300 |
0.1924279239 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_001113_030 |
0.1987245643 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 5 |
Hb_007676_020 |
0.1988216623 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001114_070 |
0.1995023434 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 7 |
Hb_124315_030 |
0.2109646373 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 8 |
Hb_031862_040 |
0.215017299 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 9 |
Hb_011716_050 |
0.2210179459 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 10 |
Hb_000608_020 |
0.2233781987 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 11 |
Hb_000685_060 |
0.2235517024 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_020805_170 |
0.2303637278 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 13 |
Hb_089032_010 |
0.2349041652 |
- |
- |
PREDICTED: flotillin-like protein 3 [Jatropha curcas] |
| 14 |
Hb_008066_070 |
0.2349609459 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 15 |
Hb_012098_170 |
0.2352825175 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |
| 16 |
Hb_006453_010 |
0.2360836032 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 17 |
Hb_116349_040 |
0.236511855 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator ARR5 [Jatropha curcas] |
| 18 |
Hb_005736_070 |
0.2385055424 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 19 |
Hb_001959_120 |
0.2403290199 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 20 |
Hb_000672_150 |
0.2416953074 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |