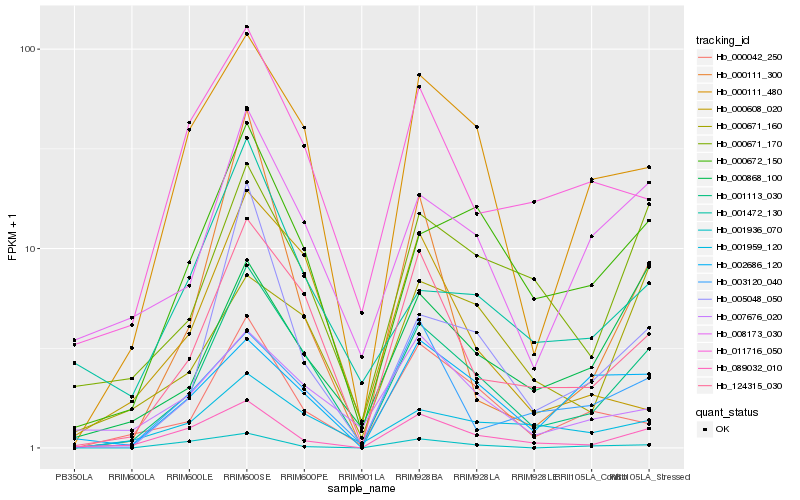
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001113_030 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 2 |
Hb_124315_030 |
0.1298226838 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 3 |
Hb_000111_480 |
0.1596470334 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 4 |
Hb_000672_150 |
0.1666168866 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 5 |
Hb_008173_030 |
0.1839435015 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 6 |
Hb_001959_120 |
0.1888366977 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 7 |
Hb_089032_010 |
0.1915674397 |
- |
- |
PREDICTED: flotillin-like protein 3 [Jatropha curcas] |
| 8 |
Hb_005048_050 |
0.192117119 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_000671_170 |
0.1943579764 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000042_250 |
0.1987245643 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000868_100 |
0.2090624686 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 12 |
Hb_011716_050 |
0.2145448217 |
- |
- |
serine acetyltransferase [Hevea brasiliensis] |
| 13 |
Hb_001472_130 |
0.2202231406 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 14 |
Hb_003120_040 |
0.2277247955 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 15 |
Hb_007676_020 |
0.2279520091 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002686_120 |
0.2327535068 |
- |
- |
short-chain type dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000111_300 |
0.233744874 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 18 |
Hb_000671_160 |
0.2350746337 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000608_020 |
0.2360393491 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 20 |
Hb_001936_070 |
0.2431923685 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM1-like [Jatropha curcas] |