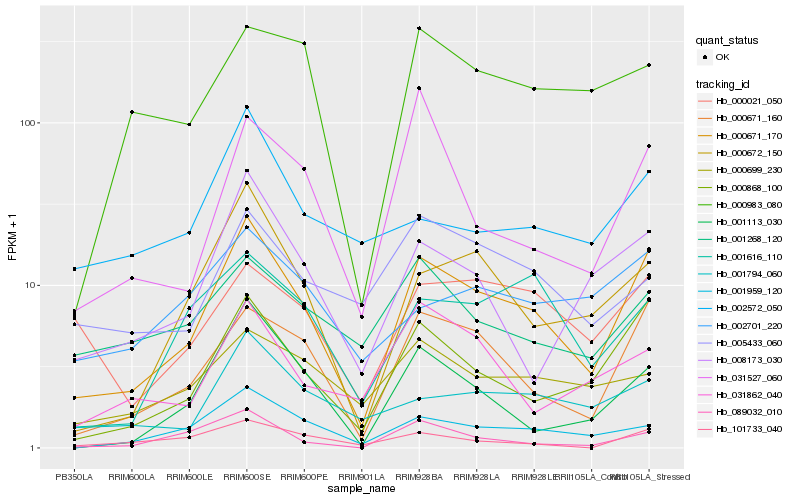
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000671_170 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_000868_100 |
0.1384998536 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 3 |
Hb_000671_160 |
0.1388915857 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000672_150 |
0.179868245 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 5 |
Hb_001959_120 |
0.1837637364 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 6 |
Hb_001113_030 |
0.1943579764 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 7 |
Hb_008173_030 |
0.1983017337 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 8 |
Hb_089032_010 |
0.2020089373 |
- |
- |
PREDICTED: flotillin-like protein 3 [Jatropha curcas] |
| 9 |
Hb_001794_060 |
0.2033787112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_101733_040 |
0.2085294598 |
- |
- |
cytochrome P450 CYP71B63v2 [Populus trichocarpa] |
| 11 |
Hb_001616_110 |
0.2085545565 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 12 |
Hb_000021_050 |
0.2133937682 |
- |
- |
PREDICTED: uncharacterized protein LOC105645238 [Jatropha curcas] |
| 13 |
Hb_031527_060 |
0.2167871416 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 14 |
Hb_000699_230 |
0.2193476133 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 15 |
Hb_031862_040 |
0.2196520042 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 16 |
Hb_001268_120 |
0.2223888107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000983_080 |
0.2225637174 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 18 |
Hb_002701_220 |
0.2240683847 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 19 |
Hb_005433_060 |
0.2256215766 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C [Jatropha curcas] |
| 20 |
Hb_002572_050 |
0.2266609104 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |