| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
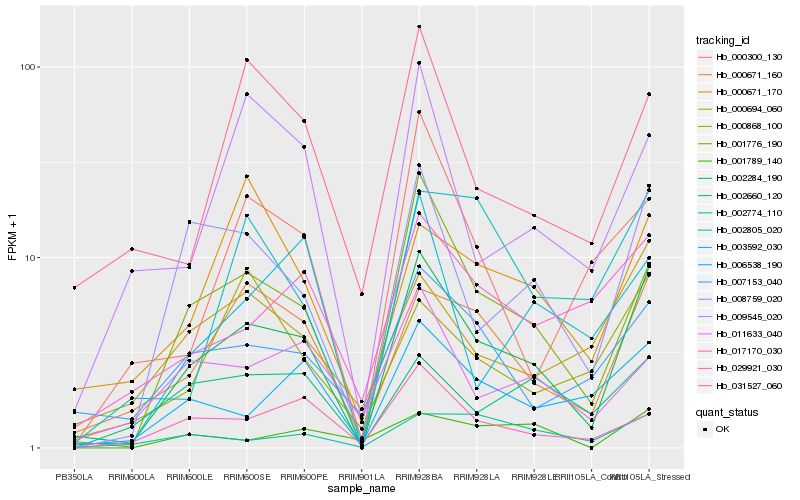
Hb_002284_190 |
0.0 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 2 |
Hb_001776_190 |
0.1575503291 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_008759_020 |
0.1834565244 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 4 |
Hb_000671_160 |
0.1912017231 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007153_040 |
0.212441685 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006538_190 |
0.2157002352 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000694_060 |
0.2253572621 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 8 |
Hb_017170_030 |
0.2268507361 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 9 |
Hb_029921_030 |
0.2330181535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_031527_060 |
0.2361008091 |
- |
- |
glutamate decarboxylase, putative [Ricinus communis] |
| 11 |
Hb_009545_020 |
0.2423796759 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |
| 12 |
Hb_011633_040 |
0.2445630084 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 13 |
Hb_003592_030 |
0.2465782361 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 14 |
Hb_002774_110 |
0.2469117972 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_002660_120 |
0.2488887462 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 16 |
Hb_002805_020 |
0.25014008 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_000300_130 |
0.2534463669 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 18 |
Hb_001789_140 |
0.2552095194 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Citrus sinensis] |
| 19 |
Hb_000671_170 |
0.256009512 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_000868_100 |
0.2566178953 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |